Trypsin and Chymotrypsin
The chymotrypsin/trypsin family of serine
proteases
Serine proteases are a large, ubiquitous family, being found in viruses, bacteria and eukaryotes. There are over twenty different subfamilies (S1-S27) based on structural and functional features. These families can be grouped into six clans (SA, SB, SC, SE, SF and SG) that differ markedly in structure, suggesting different evolutionary origins for the serine proteases. Several of these peptidases share similarities in their reaction mechanisms; chymotrypsin/trypsin, subtilisin and carboxypeptidase C subfamilies have a common catalytic triad of serine (nucleophile), aspartate (electrophile) and histidine (base). The geometric orientations of these catalytic residues are similar despite differences in protein folds. The linear order of the catalytic triad residues reflects family relationships, with the chymotrypsin/trypsin subfamily displaying the order His-Asp-Ser. The chymotrypsin/trypsin subfamily of serine proteases belongs to the Merops peptidase family S1, subfamily S1A (chymotrypsin subfamily, clan PA(S)), and is almost totally confined to animals, although trypsin-like enzymes are found in actinomycetes of the genera Streptomyces and Saccharopolyspora, and in the fungus Fusarium oxysporum. Family members mostly function extracellularly in roles that include food digestion, fibrinolysis and complement activation, but can occasionally function intracellularly, such as in the intracellular digestion of bacteria in neutrophils.
P07477 Human trypsinogen 1 entry from InterPro
|
InterPro Entry |
Method accession |
Graphical match |
Method name |
|
|
trypsin |
||
|
|
TRYPSIN_HIS |
||
|
|
TRYPSIN_SER |
||
|
|
TRYPSIN_DOM |
||
|
|
Tryp_SPc |
||
|
|
CHYMOTRYPSIN |
||
|
|
Ser_prot_trysin |
||
|
NONE |
|
1trnA1 |
|
|
NONE |
|
d1trna_ |
What InterPro tells us
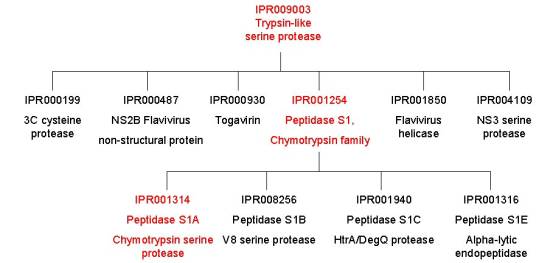
From the graphical match above, you can see that there are three InterPro entries associated with trypsin I (the same applies to the other trypsins and chymotrypsin). These different entries give you information on the family relationships of trypsin. If we start with the entry IPR009003, it has one SUPERFAMILY signature, SSF50494, representing the trypsin-like serine proteases, based on structural information. This is a large family with over 4000 protein matches. If you follow the links to IPR009003, you will find that there are several InterPro families listed under the section labelled ‘Children’, which represent the smaller, more closely related families that IPR009003 has been subdivided into. Trypsin is found within the family IPR001254, the peptidase S1, chymotrypsin family (as mentioned above there are families S1-S27). This family is represented by five signatures: PF00089 from PFAM, PS50240 profile from PROSITE and SM00020 from SMART covering the core sequence; PS00134 pattern from PROSITE covering the histidine active site and PS00135 pattern from PROSITE covering the serine active site. This family has also been subdivided into smaller families, or children: S1A, S1B, S1C and S1E; trypsin belongs to the IPR001314 peptidase S1A family. IPR001314 has one signature from PRINTS, PR00722, which was derived from three elements around the active site: the catalytic histidine, the catalytic aspartate and the catalytic serine. To give you an idea of the family relationships, look at the tree derived from IPR009003 below:
The last two entries in the table are from the structural classification databases CATH (1trnA1) and SCOP (d1trna_), the names being derived from the PDB entries that they describe (for example, d1trna_ is from PDB entry 1trn). These two entries lack an InterPro accession number, because they are structural links associated with specific proteins that have structures in PDB (here, trypsinogen 1). Following the links will take you the CATH and SCOP pages describing the structural classification of this protein.
What the structure tells us
A description and visualisation of the structural features of trypsin and chymotrypsin can be found at the PDB database. Here you can the structural details of the serine protease reaction mechanism.
Next: Table of trypsin and chymotrypin proteins
Previous: Pancreatitis, when control fails