G Proteins
What InterPro Tells Us:
Family Ties
G protein classes are defined based on the sequence and function of their a subunits, which fall into four main classes: GaS, GaQ, GaI and Ga12. There are many isoforms of each of the three subunits found in multiple combinations, some of which are found only in specialised cells. Thus far, genes encoding at least sixteen a subunits, five b subunits and 12 g subunits have been identified in humans (see Table of G Proteins). The a, b and g subunits are not related by sequence, and are therefore placed within three different InterPro families. An example of each subunit from InterPro is presented below.
P10824 Rat G protein G(I), a-1
subunit
InterPro Domain Architecture
![]()
InterPro Entry |
Method Accession |
Graphical Match |
Method Name |
|
IPR001019 |
PD000281 |
|
Gprotein_alpha
|
|
IPR001019 |
PF00503 |
|
G-alpha |
|
IPR001019 |
PR00318 |
|
GPROTEINA |
|
IPR001408 |
PR00441 |
|
GPROTEINAI |
|
IPR011025 |
SSF47895 |
|
Transducn_insert |
Classification
|
PDB Chain/Domain ID
|
PDB Chain/Structural Domains
|
|
|
1cip |
1cipa |
|
|
|
3.40.50.300.8 |
1cipA1 |
|
|
|
1.10.400.10.1 |
1cipA2 |
|
|
|
a.66.1.1 |
d1cipa1 |
|
|
|
c.37.1.8 |
d1cipa2 |
|
|
From the graphical match table above, you can see that the signatures (method accession) are divided into three InterPro entries for rat G(i) protein a subunit. These entries give information about the domain architecture of the protein, as well as its family relationships.
To look at the family relationships that involve this protein, we need to start with the entry IPR001019, which has three signatures representing the G protein a subunit family: PD000281 from the PRODOM database, PF00503 from the PFAM database, and PR00318 from the PRINTS database. This is a large family, and if you follow the links to IPR001019, you will find that there are several InterPro families listed under the section labelled ‘Children’; the ‘Children’ represent different groups of a subunits that form more closely related families based on their sequences. To find out all the family relationships within the G protein a subunit family, you can either follow the individual links to the different InterPro entries, or you can follow the link labelled ‘[tree]’ found directly underneath the ‘Children’ tag (or follow the link provided here):
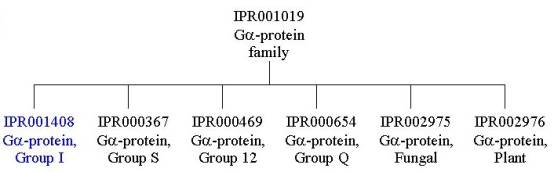
The rat G(i) protein a subunit discussed here, belongs to the Ga protein group I, as represented by InterPro entry IPR001408, which is identified by one signature, PR00441 from the PRINTS database. Group I Ga proteins include aI-like, aO-like, aT-like and aZ-like proteins. There are several other ‘Children’ of the IPR001019 G protein a subunit family.
The domain
architecture of the G protein a subunit
consists of two domains: a GTP-binding
domain homologous to Ras-like small GTPases, which includes switch regions I
and II that change conformation during activation, and a helical insertion domain unique to heterotrimeric G proteins that
sequesters the guanine nucleotide and which must be displaced to enable
nucleotide dissociation. Both these
domains are displayed in InterPro. The InterPro
entry for the (helical) insertion domain, IPR011025, has one signature, SSF47895 from the SUPERFAMILY database.
The GTP-binding domain does not have a separate entry in InterPro.
The remaining five entries in the
table above are from the structural database PDB (green stripe), and from the
structural classification databases CATH (pink stripe) and SCOP (black stripe)
(the names such as d1cipa1 are derived from the PDB entry upon which they are
based, here PDB entry 1cip, chain a, region 1). The graphical match for the PDB entry 1cipa displays the length of the
original PDB entry. The CATH and SCOP
databases give further information on the domain architecture of the rat G(i)
protein a subunit, dividing it into two
domains: the N- and C-terminal
GTP-binding domain that is structurally homologous to small GTPases (CATH 1cipA1 and SCOP d1cipa2); and the (helical) insertion
domain that interrupts the GTP-binding domain, and which is unique to
heterotrimeric G proteins (CATH 1cipA2 and SCOP d1cipa1).
P62871 (P04901) Human G protein G(I)G(S)G(T), b-1
subunit
InterPro Domain Architecture
![]()
InterPro Entry |
Method Accession |
Graphical Match |
Method Name |
|
IPR001632 |
PR00319
|
|
GPROTEINB |
|
IPR001680 |
PD000018 |
|
WD40 |
|
IPR001680 |
PF00400 |
|
WD40 |
|
IPR001680 |
PR00320 |
|
GPROTEINBRPT |
|
IPR001680 |
PS00678 |
|
WD_REPEATS_1 |
|
IPR001680 |
PS50082 |
|
WD_REPEATS_2 |
|
IPR001680 |
PS50294 |
|
WD_REPEATS_REGION |
|
IPR011046 |
SSF50978 |
|
WD40_like |
Classification |
PDB Chain/Domain ID
|
PDB Chain/Structural Domains
|
|
|
1got |
1gotb |
|
|
|
2.130.10.10.4 |
1gotB0 |
|
|
|
b.69.4.1 |
d1gotb_ |
|
|
The G protein b subunit has one domain comprised of a
beta-propeller containing WD-40 repeats with an N-terminal helix. From the graphical match table above, you can
see that the signatures are divided into three InterPro entries for this human
G protein b subunit. The InterPro entry
IPR001632 represents the G protein b subunit family, and has one signature, PR00319 from the PRINTS database. The
next six entries in the table are from IPR001680, which represents the WD-40 repeats found in G protein b subunits: PD000018 from the PRODOM database, PF00400 from the PFAM database, PR00320 from the PRINTS database, and PS00678, PS50082 and PS50294 from the PROSITE database. The
InterPro entry IPR011046 also represents the WD-40
repeats, but includes a broader classification; it has one signature, SSF50978 from the SUPERFAMILY database.
The remaining three entries
represent the original PDB entry (1gotb), and its classification in CATH
(1gotB0) and SCOP (d1gotb_).
P16874 Mouse G protein G(I)G(S)G(O), g-2
subunit
InterPro Domain Architecture
![]()
InterPro Entry |
Method Accession |
Graphical Match |
Method Name |
|
IPR001770 |
PD003783 |
|
G-gamma |
|
IPR001770 |
PF00631 |
|
G-gamma |
|
IPR001770 |
PR00321 |
|
GPROTEING |
|
IPR001770 |
PS50058 |
|
G_PROTEIN_GAMMA
|
|
IPR001770 |
SSF48670 |
|
G-gamma |
Classification
|
PDB Chain/Domain ID
|
PDB Chain/Structural Domains
|
|
|
1gp2 |
1gp2g |
|
|
|
4.10.260.10.1
|
1gp2G0 |
|
|
|
a.137.3.1
|
d1gp2g_ |
|
|
The G protein g subunit contains one domain comprised of an extended a-helical polypeptide. From the graphical match table above, you can see that all five signatures are found in one InterPro entry, IPR001770, representing the G protein g subunit. These signatures are PD003783 from the PRODOM database, PF00631 from the PFAM database, PR00321 from the PRINTS database, PS50058 from the PROSITE database, and SSF48670 from the SUPERFAMILY database. The remaining entries represent the original PDB entry (1gp2g), and its classification in CATH (1gotB0) and SCOP (d1gotb_).
What the Structure Tells Us
Structures
associated with any one of the three proteins represented above can be viewed
using AstexViewer®, which is linked from the Match Table diagrammed above via
the logo ![]() on the InterPro page (please note, there is
no link directly from this page to the AstexViewer®, therefore you need to go
to the link on the InterPro page for
Ga P10824, Gb P62871 (P04901), or Gg P16874). The AstexViewer® displays the PDB
structure with the CATH or SCOP domain highlighted (therefore each
on the InterPro page (please note, there is
no link directly from this page to the AstexViewer®, therefore you need to go
to the link on the InterPro page for
Ga P10824, Gb P62871 (P04901), or Gg P16874). The AstexViewer® displays the PDB
structure with the CATH or SCOP domain highlighted (therefore each ![]() link highlights a different region).
link highlights a different region).
There are structures available for G protein a, b and g subunits from several different organisms, including human, rat, mouse, and bovine species, amongst others, in the Protein Data Bank (PDB). A detailed description and visualisation of the structural features of G proteins can be found at the PBD ‘Molecule of the Month’. The crystallographic structures of different G proteins have provided insight into the mechanism of action of these proteins.