Ubiquitin
Ubiquitin-mediated Protein Degradation
|
|
|
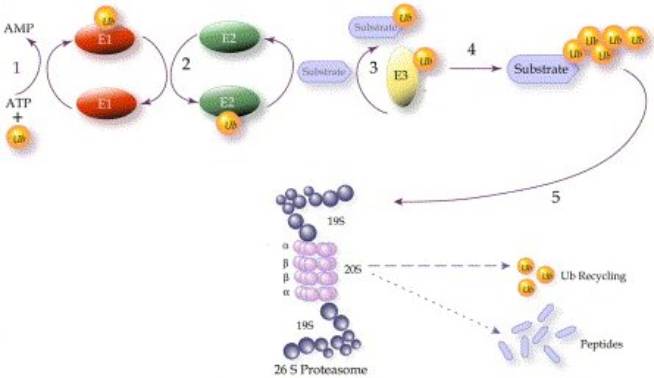
Overview of the ubiquitin-mediated protein degradation pathway. Reprinted
from European Journal of Cancer 40, A.M. Burger and A.K. Seth, The
ubiquitin-mediated protein degradation pathway in cancer: therapeutic
implications, PP.2217-2229, 2004, PMID:
15454246 |
The addition of a chain of multiple copies of ubiquitin (UB) targets a protein for destruction by the intracellular protease known as the 26S proteasome, a large complex that breaks down proteins to their constituent amino acids for reuse. The proteins targeted by this system are short-lived proteins, many of which are regulatory proteins, whose actions are controlled in part by rapid synthesis and degradation, much like an on/off switch; as such, the UB system itself is an important regulatory tool that controls the concentration of key signalling proteins. For example, many cell cycle regulatory proteins, such as cyclin, are controlled by UB-mediated proteolysis to allow a rapid transition between cell cycle stages, and to drive the direction of the cell cycle by preventing regression to an earlier stage. The selective UB-mediated degradation of proteins is also involved in the stress response, antigen processing, signal transduction, transcriptional regulation, DNA repair and apoptosis.
In addition, the 26S proteasome targets misfolded, damaged or mutant proteins with abnormal conformations that could be harmful to the cell. UB-dependent proteolysis provides the cell with a proofreading capacity for nascent polypeptide chains, whereby faulty polypeptides are targeted for destruction. Sequences that signal UB-mediated destruction can be buried in a hydrophobic core, which only becomes exposed after misfolding, providing a convenient way to distinguish misfolded proteins from functional ones - however, the presence of chaperones protects a polypeptide from degradation from the time it is synthesised until it is fully folded. Damaged proteins are also targeted. For example, hepatic cytochromes P450 are haemoproteins engaged in the oxidation of endo- and xenobiotics, during which they can become damaged by reactive intermediates; these damaged liver enzymes are rapidly removed by the UB-dependent proteolytic system.
It is important for a cell to be able to select specific proteins for degradation so as to avoid degrading proteins vital to the functioning of the cell, as well as to precisely control the delicate balance that exists between the proteins in a regulatory system, and to cope with the cell’s ever-changing protein requirements. The ubiquitin-mediated pathway achieves a high level of specificity, selecting only UB-tagged proteins to be destroyed. In addition, there exists a class of enzymes that function to remove UB from substrate proteins, thereby rescuing them from destruction by preventing indiscriminate degradation. Thus, for a protein to be degraded, it must not only have some type of UB-tagging signal, but also must escape the de-ubiquitinylation enzymes. The attachment of UB to a target protein requires the action of three enzymes, called E1 (UB-activating enzymes), E2 (UB-conjugating enzymes) and E3 (UB ligases), which work sequentially in a cascade:
Ubiquitin activation
E1 enzymes are responsible for activating UB, the first step in ubiquitinylation. The E1 enzyme hydrolyses ATP and adenylates the C-terminus of UB, and then forms a thioester bond between the C-terminus of UB and the active site cysteine of E1. To be fully active, E1 must non-covalently bind to and adenylate a second UB molecule. The E1 enzyme can then transfer the thioester-linked UB to the UB-conjugating enzyme, E2, in an ATP-dependent reaction.
Ubiquitin conjugation
UB is linked by another thioester bond to the active site cysteine of the E2 enzyme. There are several different E2 enzymes (>30 in humans), which are broadly grouped into four classes, all of which have a core catalytic domain, and some of which have short C- or N-terminal extensions that are involved in E2 localisation or in protein-protein interactions. The different E2 enzymes are able to interact with overlapping sets of E3 ligases.
Ubiquitin ligation
With the help of a third enzyme, E3 ligase, UB is transferred from the E2 enzyme to a lysine residue on a substrate protein, resulting in an isopeptide bond between the substrate lysine and the C-terminus of UB. UB ligation provides the key steps of substrate selection and UB transfer to the protein target, with the E3 ligases being responsible for substrate specificity and regulation of the ubiquitinylation process. Hundreds of putative E3 ligases have been identified, which bind to specific substrate sequences, or “degrons” (as they are targets for degradation), permitting the substrate specificity associated with this enzyme. There are at least four classes of E3 ligases: HECT-type (IPR000569), RING-type (IPR001841), PHD-type, and U-box containing (IPR003613). The E3 ligases are the only one of the 3 enzymes that is subjected to regulation, however balance in the UB system is also achieved through a set of de-ubiquitinylating isopeptidases that cleave UB off substrates.
Ubiquitin elongation
Additional UB molecules can be linked to the first one to form a poly-UB chain, which occurs through a particular type of E3 ligase sometimes referred to as a UB-elongation enzyme, or E4. There are seven lysine residues in UB that can be used to link UB molecules together, resulting in diverse structures. Poly-UB chains linked at different positions alters the destiny of the target protein to which it is added: Lys(11)-, Lys(29)- and Lys(48)-linked poly-UB chains target the protein to the proteasome for degradation, while Lys(6)- or Lys(63)-linked poly-UB chains (as well as mono-ubiquitinylation) signal reversible modifications in protein activity, location or trafficking. The length of the UB chain appears to be important as well, such as with Lys(48) poly-UB chains where its length influences its affinity for proteasomes. Therefore, E3 ligases provide the exquisite specificity in regards to which proteins should be targeted with UB, how many UB molecules are added to the target, and at what positions the poly-UB molecules are linked, thereby determining the future of the protein and the precise role it will play.
Proteasome
The 26S proteasome is a large (>60 subunits) complex with a 20S barrel-shaped proteolytic core consisting of alternating a and b subunits, and two 19S regulatory “caps” at either end (see diagram above). The 19S caps recognise, de-ubiquitinylate and unfold the target protein before it is pulled through the hollow core of the 20S catalytic centre, where it is dissembled into reusable amino acid components.
Disease
Inappropriate UB-mediated protein degradation has been implicated in a number of pathological conditions, especially neurodegenerative disorders that involve protein aggregation and inclusion body formation, such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease and ALS, where protein misfolding may play a role. Several Parkinson’s disease-causing mutations have been identified in genes encoding for UB-mediated degradation pathway proteins, such as the PARK2-encoded Parkin protein that causes autosomal recessive juvenile parkinsonism (AR-JP), and which appears to function as an E3 ligase. This degradation pathway is also implicated in certain forms of cancer as well.