Calcium pumps
P04191 Rabbit sarcoplasmic/endoplasmic reticulum calcium ATPase 1 (SERCA1) entry from InterPro:
|
Interpro Entry |
Method accession |
Graphical match |
Method name |
|
IPR000695 |
PR00120
|
|
HATPASE |
|
IPR001757 |
PR00119 |
|
CATATPASE |
|
IPR001757 |
PS00154 |
|
ATPASE_E1_E2 |
|
IPR001757 |
TIGR01494 |
|
ATPase_P-type |
|
IPR004014 |
PF00690 |
|
Cation_ATPase_N |
|
IPR005782 |
TIGR01116
|
|
ATPase-IIA1_Ca |
|
IPR005834 |
PF00702
|
|
Hydrolase |
|
IPR006068 |
PF00689 |
|
Cation_ATPase_C |
|
IPR008250 |
PF00122 |
|
E1-E2_ATPase |
|
Classification |
Domain ID |
Structural Domains |
|
|
1.20.1110.10.1 |
1eulA2 |
|
|
|
2.70.150.10.1 |
1eulA1 |
|
|
|
3.40.1110.10.1 |
1eulA3 |
|
|
|
3.40.50.1000.2 |
1eulA4 |
|
|
|
b.82.7.1 |
d1eula1 |
|
|
|
c.108.1.7 |
d1eula2 |
|
|
|
d.220.1.1 |
d1eula3
|
|
|
|
f.33.1.1
|
d1eula4
|
|
|
What InterPro tells us
From the graphical match above, you can see that the signatures in InterPro are subdivided into several groups for the sarcoplasmic/endoplasmic reticulum (SERCA) calcium pump. These groups give you information about the domain architecture of the protein, as well as its family relationships.
Starting with the domain architecture of the calcium pump, there are four major domains represented above: the transmembrane domain M, and three cytoplasmic domains A, N and P. The last eight entries in the table above are from the structural classification databases CATH and SCOP (the names such as d1eulA1 are derived from the PDB entry that they are based on, here PDB entry 1eul, chain A, region 1). If you follow the links to these databases, you will find descriptions of the structural features and classification of these domains, as well as links to the corresponding PDB entries. Domain 1eulA2 from CATH and d1eula4 from SCOP describe the transmembrane domain M, which is comprised of ten helices involving three regions of the polypeptide chain, and which contains the calcium binding sites. Domain 1eulA1 from CATH and d1eula1 from SCOP describe the cytoplasmic transduction domain A (actuator), which is interrupted by a transmembrane region (as shown in CATH), and which functions as an actuator or anchor for domain N. Domain 1eulA3 form CATH and d1eula3 form SCOP describe the cytoplasmic ATP-binding domain N (nucleotide). Domain 1eulA4 in CATH and d1eula2 in SCOP describe the cytoplasmic catalytic domain P (phosphorylation), which contains the Asp351 phosphorylation residue, around which are clustered the residues critical for ATP hydrolysis; this domain is interrupted by domain N.
There are also InterPro entries describing some of these domains. IPR004014 has one signature, PF00690 from the PFAM database, which describes the N-terminal region, while IPR006068, represented by PF00689, describes the C-terminal region. IPR008250 describes the region associated with P-type ATPases, and is represented by the signature PF00122 from the PFAM database, which covers part of the transmembrane domain and domain A.
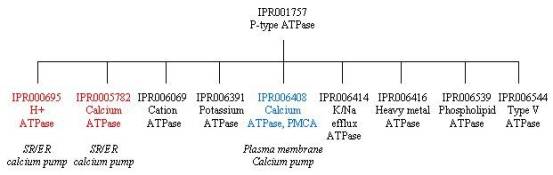
To look at the family relationships that involve the SERCA1 calcium pump, we need to start with the InterPro entry IPR001757, which is represented by three signatures: PR00119 from PRINTS, PS00154 from PROSITE (derived from residues around phosphorylated aspartate) and TIGR01494 from TIGRFAM. This is a large superfamily with over 1000 protein matches that represent P-type ATPases. This superfamily is subdivided into several smaller families based on sequence homology and function, as shown below. The SERCA1 calcium pump is found in two of those families: IPR000695 (PR00120) that represents H+ transporting ATPases, including calcium ATPases, and IPR005782 (TIGR01116) that represents calcium ATPases. By contrast, the plasma membrane calcium pump (PMCA) is represented by a different subfamily of IPR001757, namely IPR006408. The SERCA1 calcium pump is also represented by another distantly related family of proteins in IPR005834 (PF00702), which represents haloacid dehalogenase (HAD)-like hydrolases. This connection exists because the phosphorylation domain P of the calcium pump has the same Rossmann fold and the same arrangement of catalytic residues as proteins in the HAD family, implying a common catalytic mechanism; as a result, the signature for this family covers approximately the same sequence as domain P.
What the structure tells us
A description and visualisation of the structural features of the calcium pump can be found at the PDB database. The crystallographic structures of calcium-bound and unbound calcium pump have provided insights into the mechanism of calcium transport, which involves large conformational changes in all three cytoplasmic domains, as well as significant changes in several transmembrane helices.
Next: Table of calcium pump proteins
Previous: Different forms, different functions