Phenylalanine hydroxylase
By
Jennifer McDowall
To view
structure of Phenylalanine hydroxylase
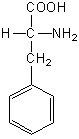
Amino acids are the building blocks of proteins, and their sequence in a protein determines the 3-dimensional shape and ultimately the function of that protein. Of the twenty amino acids commonly found in nature, nine are considered to be “essential” in humans, because they cannot be synthesised from other precursors, and therefore must be included in the diet. These essential amino acids are phenylalanine, histidine, isoleucine, leucine, lysine, methionine, threonine, tryptophan and valine. Even though these amino acids are deemed essential, their relative amounts in the bloodstream must be tightly controlled. For example, with phenylalanine (Phe), the accumulation of Phe can lead to neurological damage, while too rapid a degradation of Phe will lead to the depletion of stores of this essential amino acid and consequently a decrease in protein synthesis. Enzymes control the levels of amino acids in the bloodstream, which can fluctuate widely due to dietary intake. In the case of Phe, the degradative enzyme phenylalanine hydroxylase is the major enzyme for Phe disposal, and is responsible for serum Phe levels.
Phenylalanine hydroxylase, an
essential role
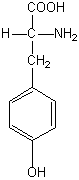
Phenylalanine hydroxylase (PheH; EC 1.14.16.1) is an iron-dependent, tetrahydrobiopterin (BH4)-dependent monooxygenase that catalyses the rate limiting step in the catabolism of Phe. PheH converts Phe into tyrosine (Tyr) by hydroxylating Phe, using BH4 as a reductant:
|
|
|
PheH |
|
|
|
L‑phenylalanine |
+ BH4 + O2 |
ŕ |
L‑tyrosine |
+ 4a‑hydroxy‑BH4 |
PheH is abundant in the liver, where it acts as a monooxygenase, incorporating one molecule of oxygen into the amino acid substrate, while the other oxygen atom is reduced to water using BH4 as the reductant.
PheH occurs in both eukaryotic and prokaryotic species. In mammals, PheH occurs as homodimers and active homotetramers in equilibrium. Each monomer polypeptide is composed of three domains: an N-terminal regulatory domain that contains an autoregulatory sequence (ARS) at the N-terminus, a central catalytic core, and a short C-terminal tetramerisation domain that contains an a-helix used to interlock the monomers. The catalytic core contains a hydrophobic “cage” that surrounds the aromatic, hydrophobic Phe substrate. The N-terminal ARS physically blocks the entrance to the active site, and may control access of both substrate and cofactors. The ARS may also interact with Phe substrate, BH4 cofactor and catecholamine inhibitors to regulate PheH activation.
In bacteria, PheH occurs as a monomer, consisting of only a single domain – the catalytic core. There is no regulatory domain present, and consequently no evidence of allosteric activation. In bacteria, Phe is not an essential amino acid, but is synthesised.
Enzyme Control
PheH controls the rate of Phe catabolism, and indirectly the rate of protein and neurotransmitter biosynthesis. In addition, the metabolites of Phe degradation are toxic to the developing brain; therefore, PheH must be under strict regulatory control in eukaryotes, despite fluctuations in dietary intake. PheH is regulated in three main ways: BH4 cofactor inhibition, substrate activation, and phosphorylation.
In addition to being an essential co-substrate for catalysis, the BH4 cofactor helps regulate the enzyme. The binding of the BH4 cofactor at the active site results in a PheH‑BH4 complex that inhibits both substrate activation by Phe and phosphorylation of the enzyme, possibly by pulling the ARS region further into the active site to block it. Furthermore, BH4 may be important for enzyme stability, shielding the active site against damage by reactive oxygen species.
Activation by its substrate, Phe, is a reversible process that involves all subunits of the tetramer. Phe can bind at two sites, the active site where it undergoes hydroxylation, and an allosteric site where it regulates PheH activation. The binding of Phe induces both local and global conformational changes in the enzyme, which displace the BH4 cofactor bound at the active site. The displacement of the BH4 cofactor releases its interaction with the ARS, which displaces the ARS from the active site as well as exposing it to phosphorylation.
Phosphorylation of PheH by cAMP-dependent protein kinase (PKA) involves a single serine residue, Ser16, in the N-terminal ARS region. Phosphorylation of Ser16 induces a conformational change in the ARS region and in the Phe-binding region of the active site. These conformational changes permit greater accessibility of Phe to the active site by displacing the ARS, as well as increasing the binding affinity of the active site for Phe. In this way, phosphorylation acts as a conformational switch that facilitates Phe-driven PheH activation. The activation of PheH is a cooperative process, where the subsequent binding of Phe serves to increase the rate of PheH phosphorylation, driving the activation of the enzyme. By contrast, the binding of the BH4 cofactor inhibits phosphorylation.
Next: Phenylketonuria, Loss of
Control