Phenylalanine hydroxylase
What InterPro Tells Us:
Family Ties
PheH belongs to the iron-dependent, BH4‑dependent aromatic amino acid hydroxylase (or monooxygenase) family that incorporate molecular oxygen into the ring of their substrates. Other family members include tyrosine hydroxylase (TyrH) and tryptophan hydroxylase (TrpH); the eukaryotic enzymes are all tetramers. TyrH is found in the central nervous system and in the adrenal gland, where it catalyses the hydroxylation of tyrosine to dihydrophenylalanine (DOPA), the first step in the biosynthesis of the catecholamine neurotransmitters. TrpH is found also found in the central nervous system, where it catalyses the hydroxylation of tryptophan to 5‑hydroxytryptophan, the first step in the biosynthesis of the neurotransmitter serotonin. In Drosophila, PheH and TyrH are encoded by the same gene.
The catalytic cores of PheH, TyrH and TrpH are homologous with a high level of both structural and sequence conservation that suggests a common catalytic mechanism, yet with different substrate specificities and some differences in the cofactor‑binding site. In particular, there are differences in the active site crevice where the substrate is thought to bind. By contrast, the N-terminal regulatory domains of these hydroxylases show very little identity, consistent with their different mechanisms of regulation. All three hydroxylases are regulated in part by phosphorylation of serine residues in the regulatory domains, which appears to convert the enzymes between active and inactive forms. However, PheH is also regulated by its substrate and by the BH4 cofactor.
The N-terminal regulatory domain has a similar fold to the ‘ACT domain’, a conserved regulatory ligand-binding fold found in Aspartate kinase, Chorismate mutase and TyrA (prephenate dehydrogenase – a key enzyme in bacterial Phe biosynthesis), although the PheH ACT-like domain has a different ligand-binding mode. The ACT domain fold is ferredoxin-like, containing a double bab motif (babbab). The ACT domain consensus sequence has been detected in the sequences of several proteins involved in amino acid and purine metabolism. These domains are regulated by small-molecule effectors.
P00439 Human Phenylalanine
Hydroxylase
InterPro Domain Architecture
![]()
|
InterPro Entry |
Method Accession |
Graphical Match |
Method Name |
|
IPR001273 |
PD002559 |
|
Aaa_hydroxylase |
|
IPR001273 |
PF00351 |
|
Biopterin_H |
|
IPR001273 |
PR00372 |
|
FYWHYDRXLASE |
|
IPR001273 |
PS00367 |
|
BIOPTERIN_HYDROXYL |
|
IPR002912 |
PF01842 |
|
ACT |
|
IPR005961 |
TIGR01268 |
|
Phe4hydrox_tetr |
|
Classification |
PDB Chain/Domain ID |
PDB Chain/Structural Domains |
|
|
1j8u |
1j8ua |
|
|
|
2pah |
2paha |
|
|
|
2pah |
2pahb |
|
|
|
1.10.800.10.1 |
1j8uA0 |
|
|
|
d.178.1.1 |
d1j8ua_ |
|
|
From the graphical match above, you can see that the signatures (method accession) are divided into three InterPro entries for human phenylalanine hydroxylase. These entries give information about the domain architecture of the protein, as well as its family relationships
To look at the family relationships that involve this protein, we need to start with entry IPR001273, which has four signatures representing the aromatic amino acid hydroxylase family: PD002559 from the PRODOM database, PF00351 from the PFAM database, PR00372 from the PRINTS database, and PS00367 from the PROSITE database. These signatures are based on the catalytic domain, which is highly conserved in this family. This family includes the tyrosine hydroxylase and tryptophan hydroxylase enzymes, to which phenylalanine hydroxylase is related. If you follow the links to IPR001273, you will find that there are several InterPro families listed under the section labelled ‘Children’; the ‘Children’ represent different groups of aromatic amino acid hydroxylases that form more closely related families based on sequence and function. To find all the family relationships within the aromatic amino acid hydroxylase family, you can either follow the individual links to the different InterPro entries, or you can follow the link labelled ‘[tree]’ found directly underneath the ‘Children’ tag (or follow the link provided here).
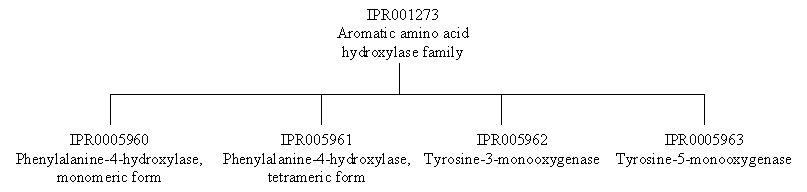
Human phenylalanine hydroxylase belongs to IPR005961, the phenylalanine-4-hydroxylase (tetrameric form) family, which is represented by one signature, TIGR01268 from the TIGR database (IPR005960 for the monomeric form is represents the bacterial enzymes). This signature is based on the full length of the protein.
The domain architecture of human phenylalanine hydroxylase consists of three domains: the N-terminal regulatory domain, the central catalytic domain and the C-terminal tetramerisation domain. IPR002912 represents the amino acid–binding ACT domain, which covers the N-terminal regulatory domain in phenylalanine hydroxylase. If you follow the links to this entry, you will find many entries under the section labelled ‘Found in’, which represents the list of protein families that contain this domain. This entry is represented by one signature, PF01842 from the PFAM database.
The remaining five entries in the table above are from the structural database PDB (green stripe), and from the structural classification databases CATH (pink stripe) and SCOP (black stripe) (the names such as 1j8uA0 are derived from the PDB entry upon which they are based, here PDB entry 1j8u, chain A). The graphical match for the PDB entry 1j8ua displays the length of the original PDB entry, here covering the catalytic domain. The CATH (1j8uA0) and SCOP (d1j8ua_) give information on the classification of the PDB structure for the catalytic domain.
What the Structure Tells Us
Structures
associated with phenylalanine hydroxylase can be viewed using AstexViewer®,
which is linked from the Match Table above via the logo ![]() on the InterPro page (please note, there is no link directly from
this page to the AstexViewer®, therefore you need to go to the link on the
InterPro page for P00439). The AstexViewer® displays the PDB structure with the CATH or SCOP
domain highlighted.
on the InterPro page (please note, there is no link directly from
this page to the AstexViewer®, therefore you need to go to the link on the
InterPro page for P00439). The AstexViewer® displays the PDB structure with the CATH or SCOP
domain highlighted.
There are structures available for phenylalanine hydroxylase from several different species in the Protein Data Bank (PDB). A detailed description and visualisation of the structural features of phenylalanine hydroxylase can be found at the PDB ‘Molecule of the Month’. The crystallographic structures of different phenylalanine hydroxylases have provided insight into the mechanism of action of these enzymes.
