Acetylcholine Receptors
What InterPro Tells Us
An example of a muscarinic and a nicotinic AchR is given below.
P08172 Human Muscarinic Acetylcholine Receptor M2
InterPro Domain Architecture
![]()
InterPro Entry |
Signatures |
Graphical Match |
Method Name |
|
IPR000276 |
PF00001 |
|
7tm_1 |
|
IPR000276 |
PR00237 |
|
GPCRRHODOPSN |
|
IPR000276 |
PS00237 |
|
G_PROTEIN_RECEP_F1_1 |
|
IPR000276 |
PS50262 |
|
G_PROTEIN_RECEP_F1_2 |
|
IPR000995 |
PR00243 |
|
MUSCARINICR |
|
IPR001065 |
PR00539 |
|
MUSCRINICM2R |
|
IPR001065 |
PTHR19266:SF84 |
|
M2_receptor |
|
Structural Predictions |
|
|
|
|
MB_P08172 |
|
|
|
From the graphical match above,
you can see that the signatures are grouped into three InterPro entries for the
human muscarinic acetylcholine receptor M2.
These entries give information about the family relationships involving
this protein. Starting at the top of
the hierarchy is IPR000276, which represents the
Rhodopsin-like G-protein-coupled receptor family. This is a very large family with over 12,000 protein members that
include not only the muscarinic AChRs, but over seventy other receptor families
as well, such as melatonin receptors, dopamine receptors, interleukin receptors,
histamine receptors and opioid receptors, to name but a few. Four signatures represent PR000276: PF00001 from the PFAM database, PR00237 from the PRINTS database, and PS00237 and PS50262 from the PROSITE database, where the PS00237 signature is derived
from the conserved triplet that spans the major part of the third transmembrane
helix. If you follow the links to IPR000276,
you will find that the different rhodopsin-like GPCR families are listed under
the section labelled ‘Children’; the ‘Children’ represent different
groups receptors that form more closely related families based on sequence and
function. The InterPro entry IPR000995 is a ‘Child’ of IPR000276, and
represents the muscarinic acetylcholine receptor family. IPR000995 has one signature, PR00243 from the PRINTS database.
This family includes all the different receptor subtypes, M1-M5, each of
which is represented by its own entry as ‘Children’ of IPR000995. The Child represented here is IPR001065,
representing the muscarinic acetylcholine receptor M2 family, which has two
signatures: PR00539 from the PRINTS database and PTHR19266:SF84 from the PANTHER database.
The remaining entry in the table above gives information on the predicted structure of this protein, and is from the homology model database ModBase (yellow stripe), MB_P08172, which provides a possible structure for the M2 muscarinic AchR.
P25110 Rat Nicotinic Acetylcholine
Receptor, Delta Subunit
InterPro Domain Architecture
![]()
InterPro Entry |
Signatures |
Graphical Match |
Method Name |
|
IPR002394 |
PR00254 |
|
NICOTINICR |
|
IPR006029 |
PF02932 |
|
Neur_chan_memb |
|
IPR006201 |
PR00252 |
|
NRIONCHANNEL |
|
IPR006201 |
PS00236 |
|
NEUROTR_ION_CHANNEL |
|
IPR006201 |
PTHR18945 |
|
Neur_channel |
|
IPR006201 |
TIGR00860 |
|
LIC |
|
IPR006202 |
PF02931 |
|
Neur_chan_LBD |
|
IPR006202 |
SSF63712 |
|
Neur_chan_LBD |
|
Structural Features |
PDB Chain/Domain ID |
|
|
|
1cek |
1cekA |
|
|
|
j.35.1.1 |
d1a11__ |
|
|
|
Structural Predictions |
|
|
|
|
MB_P25110 |
|
|
|
From the graphical match above, you can see that the signatures are grouped into four InterPro entries for the delta subunit of rat nicotinic acetylcholine receptor. These entries give information about the domain architecture of the protein, as well as its family relationships.
To look at the family relationships that involve this protein, we need to start with entry IPR006201, which is top of the hierarchy and has four signatures representing the neurotransmitter-gated ion-channel family: PR00252 from the PRINTS database, PS00236 from the PROSITE database, PTHR18945 from the PANTHER database, and TIGR00860 from the TIGRFAMs database. This family includes all neurotransmitter-gated ion channel proteins, including not only nicotinic AChRs, but also GABA-A receptors, Glycine receptors and 5-HT3 receptors. If you follow the links to IPR006201, you will find that these different receptor families are listed under the section labelled ‘Children’. The InterPro entry IPR002394 is a ‘Child’ of IPR006201, and represents the nicotinic acetylcholine receptor family, each member being closely related in sequence and function, which includes the alpha, beta, delta, epsilon and gamma subunits. IPR002394 has one signature, PR00254 from the PRINTS database. To see all the neurotransmitter-gated ion channel receptors that are related to nicotinic AchR, either follow the individual links to the different InterPro entries under ‘Children’ in IPR006201, or follow the link labelled ‘[tree]’ found directly underneath the ‘Children’ tag (or follow the link provided here).
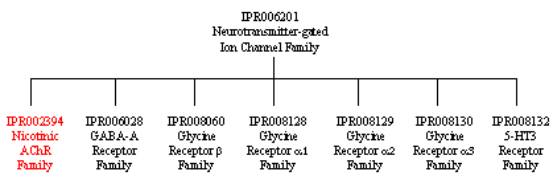
The domain architecture of the delta
subunit of rat nicotinic AChR consists of two domains: the N-terminal ligand-binding
domain and the transmembrane domain. IPR006202 represents the N-terminal ligand-binding domain of
neurotransmitter-gated ion channel proteins.
If you follow the links to this entry, you will find many entries under
the section labelled ‘Found in’, which represents the list of receptor protein
families that contain this domain. Two
signatures represent this entry: PF02931 from the PFAM database, and SSF63712 from the SUPERFAMILY database. The other domain is IPR006029, which represents the C-terminal transmembrane domain of
neurotransmitter-gated ion channel proteins.
This domain is found in many receptor protein families besides nicotinic
AchR proteins, and is represented by one signature: PF02932 from the PFAM database.
The remaining three entries in the table above give information on the structure of this protein, presenting known structural data from the structural database PDB (green stripe) and the structural classification database SCOP (black stripe) (the names such as d1a11_ are derived from the PDB entry upon which they are based, here PDB entry 1a11). The graphical match for the PDB entry 1cekA displays the length of the original PDB entry, here covering the channel-lining segment of the transmembrane domain. SCOP (d1a11__) gives information on the classification of the PDB structure for the channel-lining segment. There is also a predicted structure for this protein from the homology model database ModBase (yellow stripe), MB_P25110, which provides a possible structure for the ligand-binding domain of this protein.
What the Structure Tells Us
Structures
associated with acetylcholine receptors can be viewed using AstexViewer®, which
is linked from the Match Table via the logo ![]() on the InterPro page (please note, there is no link directly from
this page to the AstexViewer®, therefore you need to go to the link on the
InterPro page for P25110). The AstexViewer® displays the PDB structure with the SCOP domain
highlighted.
on the InterPro page (please note, there is no link directly from
this page to the AstexViewer®, therefore you need to go to the link on the
InterPro page for P25110). The AstexViewer® displays the PDB structure with the SCOP domain
highlighted.
There are
structures available for various acetylcholine receptors from several different
species in the Protein Data Bank (PDB).
A detailed description and visualisation of the structural features of acetylcholine
receptors can be found at the PDB ‘Molecule of the Month’.
The crystallographic structures of different acetylcholine receptors
have provided insight into the mechanism of action of these important receptors.
Next: Table of Acetylcholine
Receptors
Previous: Two Types of Receptors