Protein of the Month |
December 2006
MORE ON THIS MONTH’S PROTEIN
|
|
OTHER PROTEINS OF INTEREST |
|
Molecule of the Month: Transposase |
|
|
Transposase
By Jennifer McDowall
Transposition, the movement of genes from one position on the chromosome to another, was first suggested by Barbara McClintock in her 1948 publication, following years of work on chromosomal breakage and recombination. She observed that mutable loci affecting maize seed colour variegation involved chromosomal breakage and fusion, as well as the transposition of genes. She suggested that such genetic instability could occur in all organisms. Although such ‘moveable’ genes were first met with scepticism, transposable elements are now known to exist in all organisms, and she was to receive a Nobel Prize for her work in 1983.
Transposase,
Moving Genetic Elements About
In maize, McClintock originally described two ‘moveable’ elements: activator (Ac) and dissociation (Ds). Ds (non-autonomous) was found to only move in the presence of Ac, whereas Ac (autonomous) could move on its own. Either element was able to insert into different locations in the genome, often within genes, which frequently resulted in mutations in those genes. We now know that the Ds element is a deletion mutant of Ac that has lost the necessary sequences to move on its own. Other transposable elements can exist as autonomous (encode products for transposition) or non-autonomous elements.
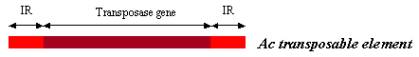
Ac is just over 4.5kb long, and encodes for a 3.5kb mRNA that is translated into a transposase protein (which is partly deleted in Ds). The transposase is responsible for excising the transposable element anywhere in the genome through its recognition of specific sequences. In Ac, the transposase gene is flanked on either side by 10bp inverted repeat sequences (IR), which are essential for transposition. Each IR contains an AAACGG motif that acts as the transposase binding sequence – mutations in this sequence would abolish the transposable element’s ability to move.
|
|
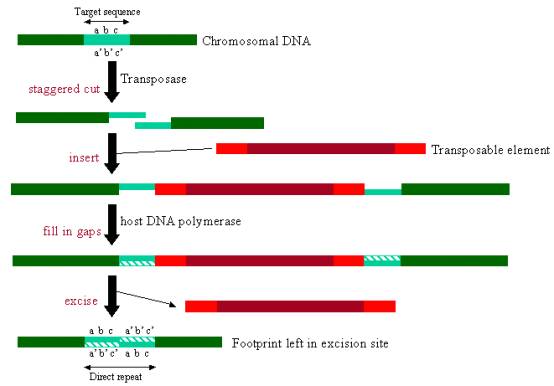
The transposase also recognises sequences within the genome, making 8bp staggered cuts within the chromosome for the transposable element to insert (DNA hairpin intermediates form in host DNA during transposition). As the cut is staggered, the 8bp single stranded genomic DNA on either side of the transposable element needs to be filled in using host DNA polymerase, thereby creating 8bp target site duplications that flank the transposable element. When the transposable element later excises from the site, the 8bp duplication remains at the site as a direct repeat to mark where the element has been as a footprint (see figure below).
|
|
|
Transposition: transposable element inserts into staggered cuts in chromosomal DNA, the gaps being filled in by DNA polymerase. Upon excision, direct repeats are left in the chromosomal DNA. |
Harnessing
Transposition
Transposable elements can insert within genes or regulatory sequences, which can disrupt gene function or expression, sometimes only temporarily if the element later excises. However, if a deleterious footprint is left by the excised element, it can permanently alter gene function or expression, sometimes even creating new alleles of a gene.
Transposable elements can also ‘pick up’ endogenous genes, moving them to different genomic sites and increasing their copy number. This is common with certain bacterial transposons, which can carry antibiotic resistance genes, such as the ampicillin-resistance transposon Tn3, kanamycin-resistance transposon Tn5, and the chloramphenicol resistance transposon Tn9.
Scientist have made use of such ability, manipulating both prokaryotic and eukaryotic transposons for use as shuttles to moving genetic material or to induce genetic mutation for experimentation.
Next: Types of Transposable Elements