Transposase
What InterPro Tells Us
P07636
Bacteriophage Mu Transposase
InterPro Domain Architecture
![]()
InterPro Entry |
Signatures |
Graphical Match |
Method Name |
|
IPR003314 |
PF02316 |
|
Mu_DNA_bind |
|
IPR011991 |
G3DSA:1.10.10.10 |
|
Wing_hlx_DNA_bd |
|
IPR009061 |
SSF46955 |
|
Putativ_DNA_bind |
|
IPR012287 |
G3DSA:1.10.10.60 |
|
Homeodomain-rel |
|
IPR009057 |
SSF46689 |
|
Homeodomain_like |
|
IPR004189 |
PF02914 |
|
Mu_transposase |
|
IPR012337 |
SSF53098 |
|
RNaseH_fold |
|
IPR009004 |
G3DSA:2.30.30.130 |
|
mu_transposase_C |
|
Structural Features |
|
|
|
|
1tns |
1tns |
|
|
|
2ezk |
2ezk |
|
|
|
2ezh |
2ezh |
|
|
|
1bco |
1bco |
|
|
|
1.10.10.10.101 |
1tns00 |
|
|
|
1.10.10.60.48 |
2ezk00 |
|
|
|
1.10.10.60.42 |
2ezi00 |
|
|
|
3.30.420.10.20 |
1bco01 |
|
|
|
2.30.30.130.1 |
1bco02 |
|
|
|
a.6.1.7 |
d1tns__ |
|
|
|
a.4.1.2 |
d2ezk__ |
|
|
|
a.4.1.2 |
d2ezh__ |
|
|
|
c.55.3.3 |
d1bco_2 |
|
|
|
b.48.1.1 |
d1bco_1 |
|
|
From the graphical match above, you can see that the signatures are all grouped into eight InterPro entries for bacteriophage Mu transposase. InterPro entries group together all the signatures that represent the same sequence found in the same set of proteins. These entries provide a hierarchical classification of the domains within Mu transposase.
DOMAIN Entries
Ø IPR003314: DNA-binding domain of MuA transposase/repressor protein CI, represented by one signature: PF02316 (PFAM).
Ø IPR011991: DNA-binding domain of winged helix repressors, represented by one signature: G3DSA:1.10.10.10 (Gene3D).
Ø IPR009061: DNA-binding domain, represented by one signature: SSF46955 (SUPERFAMILY).
Ø IPR012287: Homeodomain-related, represented by one signature: G3DSA:1.10.10.60 (Gene3D).
Ø IPR009057: Homeodomain-like, represented by one signature: SSF46689 (SUPERFAMILY).
Ø IPR004189: Bacteriophage Mu transposase core domain, represented by one signature: PF02914 (PFAM).
Ø IPR012337: RibonucleaseH-type fold domain of polynucleotidyl transferases, represented by one signature: SSF53098 (SUPERFAMILY).
Ø IPR009004: C-terminal domain of Mu transposase, represented by one signature: G3DSA:2.30.30.130 (Gene3D).
The graphical match shows that Mu transposase can be divided into five domains, all of which have a structure deposited in PDB, and which are shown under ‘Structural Features’ in the table above. The PDB structures (green stripe) have been classified by both the CATH (pink stripe) and the SCOP (black stripe) databases (the names such as 1bco02 are derived from the PDB entry upon which they are based, here PDB entry 1bco, (chain 0 as only one chain), fragment 2). Both the CATH and SCOP databases subdivide Mu transposase into five domains, similar to the InterPro signatures:
1) Winged-helix-type DNA-binding domain: winged-helix type 3-helical fold (but distinct)
2) Homeodomain-type DNA-binding domain: homeodomain-type 3-helical bundle
3) Homeodomain-type domain: homeodomain-type 3-helical bundle
4) Enzyme core: ribonuclease H-type motif of 3 layers alpha/beta/alpha
5) C-terminal domain: beta-barrel with a Greek key topology
IPR003314 covers both domains 1 and 2, as both appear to be involved in DNA-binding, however they differ in terms of their structure, domain 1 having a winged helix-type fold (IPR011991 and IPR009061), while domain 2 having a homeodomain-type fold (IPR012287). IPR009057 covers both domains 2 and 3, because they both consist of a homeodomain-type structural fold. Domain 4 forms the enzyme core (IPR004189), which has a ribonuclease H-type fold (IPR012337). Domain 5 forms the C-terminal region.
What the Structure Tells Us
Structures
associated with Mu transposase can be viewed using AstexViewer®, which is
linked from the Match Table via the logo ![]() on the InterPro page (please note, there is no
link directly from this page to the AstexViewer®, therefore you need to go to
the
on the InterPro page (please note, there is no
link directly from this page to the AstexViewer®, therefore you need to go to
the ![]() link on the InterPro page for P07636). The AstexViewer® displays the PDB structure for each of the five
domains (domains 1, 2 and 3 separately, domains 4 and 5 together), with domains
4 and 5 shown below.
link on the InterPro page for P07636). The AstexViewer® displays the PDB structure for each of the five
domains (domains 1, 2 and 3 separately, domains 4 and 5 together), with domains
4 and 5 shown below.
|
|
|
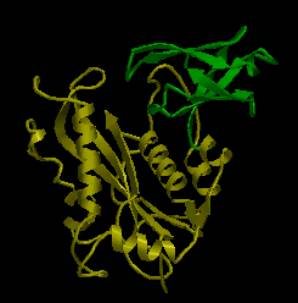
AstexView of bacteriophage Mu transposase, domains 4 and 5: domain 4 (ribonuclease H-like) is highlighted in yellow, domain 5 (beta-barrel) in green. |
There are
structures available for various transposases and integrases from various
organisms in the Protein Data Bank (PDB).
A detailed description and visualisation of the structural features of
transposase proteins can be found at the PDB ‘Molecule of the Month’.
The crystallographic structures of various transposase proteins have
provided insight into their mode of action.
Next: Table of Transposases
Previous: Types of
Transposable Elements