Clathrin
What InterPro Tells Us
P49951 Bovine clathrin heavy chain
InterPro Domain Architecture
![]()
InterPro Entry |
Signatures |
Graphical Match |
|
IPR000547 |
PF00637 |
|
|
IPR000547 |
SM00299 |
|
|
IPR001473 |
PF01394 |
|
|
IPR011990 |
G3DSA:1.25.40.10 |
|
|
IPR015348 |
PF09268 |
|
|
Structural Features |
|
|
|
|
1b89A |
|
|
|
1utcA, 1utcB |
|
|
1.25.40.10.18 |
1b89A00 |
|
|
2.130.10.110.1 |
1utcA00 |
|
|
a.118.1.3 |
d1b89a_ |
|
|
a.118.1.4 |
d1utca1 |
|
|
b.69.6.1 |
d1utca2 |
|
|
Structural Predictions |
|
|
|
|
MB_P49951 |
|
|
|
SW_P49951 |
|
From the graphical match in the table above, you can see that the signatures are grouped into four InterPro entries for bovine clathrin heavy chain. These entries identify the domain architecture and sequence features of this protein.
DOMAIN Entries
Ø IPR001473: Clathrin propeller, N-terminal, represented by one signature: PF01394 (PFAM).
Ø IPR011990: Tetratricopeptide-like helical, represented by one signature: G3DSA:1.25.40.10 (Gene3D).
Ø IPR015348: Clathrin heavy chain, linker, core motif, represented by one signature: PF09268 (PFAM).
REPEAT Entries
Ø IPR000547: Clathrin heavy chain, 7-fold repeat, represented by two signatures: PF00637 (PFAM) and SM00299 (SMART).
The bovine clathrin heavy chain consists of different repetitive sequence elements. The N-terminal is composed of seven 4-stranded beta-sheet motifs that form a beta-propeller (IPR001473). The beta-propeller is at the tip of each leg of the triskelion, and is responsible for binding clathrin adaptors and membrane attachment proteins. The linker region (IPR015348) is a short alpha-helical sequence (ARM-repeat-like) linking the N- and C-terminal domains. The C-terminal is composed of a 7-fold tetratricopeptide repeat (alpha-alpha-superhelical ARM-repeat) with a similar structure to the short linker region. The C-terminal forms the central hub of the triskelion.
Structural features
The remaining ten entries in the table above give information on the known and predicted structure of this protein. These entries present known structural data from the structural database PDB (green stripe) and the structural classification databases CATH (pink stripe) and SCOP (black stripe) (the names such as 1b89A00 are derived from the PDB entry upon which they are based, here PDB entry 1b89, chain A), as well as predicted structural data from the homology model databases ModBase (yellow stripe) and Swiss-Model (red stripe).
The graphical matches for the PDB entries 1utc and 1b89
display the full-length original PDB entries for the N- and C-terminal regions,
respectively. Both CATH (1b89A00) and SCOP (d1b89a_) classify the C-terminal region
as an ARM repeat alpha-alpha-superhelical structure, but they differ in their
classification of the N-terminal region.
CATH (1utcA00) classifies the entire N-terminal structure as an beta-propeller,
while SCOP (d1utca1; d1utca2) divides it into an N-terminal beta-propeller, and a short ARM
repeat alpha-helical linker.
The homology models from both ModBase (MB_P49951)
and Swiss-Model (SW_P49951)
cover a larger portion of the N-terminal region of the clathrin heavy chain.
What the Structure Tells Us
Structures
associated with the bovine clathrin heavy chain can be viewed using
AstexViewer®, which is linked from the Match Table via the logo ![]() on the InterPro page (please note, there is no
link directly from this page to the AstexViewer®, therefore you need to go to
the
on the InterPro page (please note, there is no
link directly from this page to the AstexViewer®, therefore you need to go to
the ![]() link on the InterPro page for P49951).
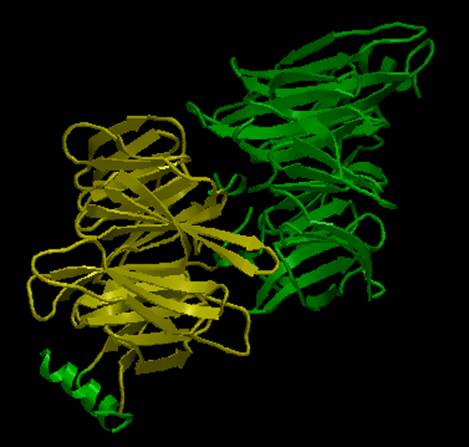
The AstexViewer® displays the PDB structure with the CATH or SCOP domain
highlighted in yellow. In the picture below,
the SCOP domain representing the N-terminal region is shown. Note that there are two copies of the
N-terminal (i.e. shown as a dimer); the beta-sheet structure in yellow
represents the N-terminal beta-propeller, while the short helix in green to its
left represents the short helical linker region (the entire N-terminal
structure is repeated in green on the right).
link on the InterPro page for P49951).
The AstexViewer® displays the PDB structure with the CATH or SCOP domain
highlighted in yellow. In the picture below,
the SCOP domain representing the N-terminal region is shown. Note that there are two copies of the
N-terminal (i.e. shown as a dimer); the beta-sheet structure in yellow
represents the N-terminal beta-propeller, while the short helix in green to its
left represents the short helical linker region (the entire N-terminal
structure is repeated in green on the right).
|
|
|
AstexView of bovine clathrin heavy chain, N-terminal region: the beta-propeller is shown in yellow, while the short alpha-helical linker is to its left in green; the region is duplicated in green on the right (i.e. shown as a dimer). |
There are structures available for
several clathrin heavy chains from different organisms in the Protein Data Bank
(PDB). A detailed description and
visualisation of the structural features of clathrin can be found at the PDB ‘Molecule of the Month’.
The crystallographic structures of clathrin lattices have provided
insight into clathrin-mediated protein transport.