Aconitase
Iron
Regulatory Protein 1, How It Works
Iron is indispensable for the functioning of many prosthetic groups, including haem and iron-sulphur clusters, and its depletion can cause anaemia. However, excess iron can catalyse the formation of reactive oxygen species that damage proteins, nucleic acids and lipids, and is associated with both haemochromatosis and thalassaemia. Iron regulatory proteins (IRPs) register cytosolic iron concentrations and post-transcriptionally regulate the expression of iron metabolism genes in order to optimize cellular iron availability. IRPs control iron metabolism by binding to specific non-coding sequences within an mRNA, known as iron-responsive elements (IRE). IREs are 30 nucleotide long RNA motifs that form special stem-loop structures. IREs occur in either the 3’-UTR (untranslated region) or 5’-UTR of an mRNA. Transcripts containing IREs include those encoding:
· Ferritin subunits L and H (expressed in liver and heart, respectively), which mediate iron storage.
· Transferrin, which binds extracellular iron and circulates it in blood plasma.
· Transferrin receptor (expressed on plasma membrane), which controls iron uptake into a cell by binding to iron-bound transferrin.
· Ferroportin, and iron exporter expressed on the surface of gut enterocytes, macrophages and liver cells.
· Divalent metal transporter 1 (DMT1), a ferrous iron transporter that functions in intestinal iron absorption.
· Mitochondrial aconitase
· Succinate dehydrogenase
· Erythroid aminolevulinic acid synthetase, which catalyses the first step in tetrapyrrole synthesis.
· Alzheimer's amyloid precursor protein.
Under conditions of iron depletion, IRP1 stabilises transferrin receptor mRNA to prolong its half-life, while inhibiting the translation of ferritin mRNA. This both enhances iron uptake into cells and stops it being sequestered away for storage. IRP1 can therefore act as both a translational enhancer and inhibitor - which way it behaves depends upon where the IRE sequence(s) are found in the transcript.
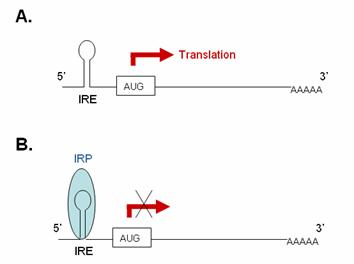
Inhibiting translation: IRP binding IRE within 5’-UTR of mRNA
In iron-deficient cells, the binding of an IRP to an IRE sequence found in the 5’UTR of an mRNA prevents its translation by blocking the mRNA from binding to the ribosome. In iron-replete cells, the absence of IRP binding allows these transcripts to be freely translated. Transcripts that carry IRE in their 5’-UTR include ferritin H and L subunits and aminolevulinic acid synthetase.
|
|
|
5’IRE: (A) In the absence of IRP these transcripts
are freely translated. (B) Binding of
IRP1 to the 5’IRE of a transcript blocks
its translation. |
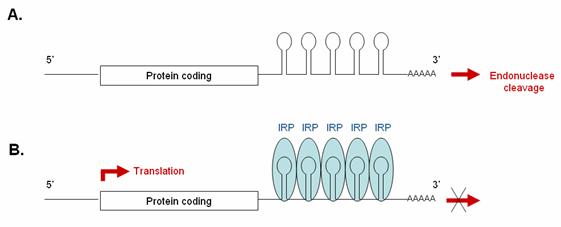
Promoting translation: IRP binding IREs within 3’-UTR of mRNA
Some transcripts contain one or more IRE sequences in their 3’UTR. In iron-deficient cells, the binding of IRPs to these 3’ IRE sequences protects them from endonuclease cleavage by blocking nuclease attack, thereby prolonging their half-life and enabling them to be freely translated. In iron-replete cells, the absence of IRP binding makes these transcripts susceptible to endonuclease attack and subsequent rapid degradation. Transcripts carrying IREs in their 3’-UTR include the transferrin receptor (5 copies) and the DMT1 transporter.
|
|
|
3’IRE: (A) In the absence of IRP, these
transcripts are rapidly degraded. (B) Binding
of IRP1 to the 3’IRE of a transcript protects it from endonucleolytic
degradation, thereby enabling it to be translated. |
Next: The
Aconitase - IRP1 Switch
Previous: Aconitase,
a moonlighting enzyme