Fatty Acid Synthase
What InterPro Tells Us
P49327 Human Fatty acid synthase
InterPro Domain Architecture
![]()
InterPro Entry |
Signatures |
Graphical Match |
|
IPR014030 |
PF00109 |
|
|
IPR014030 |
PS00606 |
|
|
IPR014031 |
PF02801 |
|
|
IPR016038 |
G3DSA:3.40.47.10 |
|
|
IPR016039 |
SSF53901 |
|
|
IPR014043 |
PF00698 |
|
|
IPR001227 |
G3DSA:3.40.366.10 |
|
|
IPR016035 |
SSF52151 |
|
|
IPR016036 |
|
|
|
IPR013217 |
PF08242 |
|
|
IPR011032 |
SSF50129 |
|
|
IPR013149 |
PF00107 |
|
|
IPR002198 |
PF00106 |
|
|
IPR016040 |
G3DSA:3.40.50.720 |
|
|
IPR016040 |
SSF51735 |
|
|
IPR006162 |
PS00012 |
|
|
IPR006163 |
PF00550 |
|
|
IPR006163 |
PS50075 |
|
|
IPR009081 |
G3DSA:1.10.1200.10 |
|
|
IPR009081 |
SSF47336 |
|
|
PF00975 |
|
|
|
IPR000794 |
PTHR11712 |
|
|
Structural Features |
|
|
|
PDB |
2jfd (chains A-D) |
|
|
PDB |
2cq5 (chain B) |
|
|
PDB |
1xkt (chains A, B) |
|
|
CATH |
1xktA02 |
|
|
CATH |
1xktA01 |
|
|
SCOP |
d1xkta |
|
|
Structural Predictions |
|
|
|
ModBase |
MB_P49327 |
|
From the graphical match in the table above, you can see that the signatures are grouped into thirteen InterPro entries for human fatty acid synthase. InterPro entries group together all the signatures that represent the same sequence found in the same set of proteins. These entries provide a family hierarchy of fatty acid synthase proteins, as well as identifying the domain architecture and sequence features of this protein.
FAMILY Entries
Ø IPR000794: Beta-ketoacyl synthase, represented by one signature: PTHR11712 (PANTHER).
This entry places FAS enzymes in the same family as several ketoacyl synthases and polyketide synthases.
DOMAIN Entries
1)
Beta-ketoacyl synthase domain (KS; EC 2.3.1.41):
Ø IPR014030: N-terminal subdomain, represented by two signatures: PF00109 (PFAM), PS00606 (PROSITE).
Ø IPR014031: C-terminal subdomain, represented by one signature: PF02801 (PFAM).
Ø IPR016038: Thiolase-like domain subgroup, represented by one signature: G3DSA:3.40.47.10 (GENE3D).
Ø IPR016039: Thiolase-like domain, represented by one signature: SSF53901 (SUPERFAMILY).
2)
Malonyl transacylase (MPT; EC 2.3.1.39) and possible dehydratase (DH; EC 4.2.1.61)
Ø IPR014043: Acyl/malonyl transferases domain, represented by one signature: PF00698 (PFAM).
Ø IPR001227: Acyl/malonyl transferases region, represented by one signature: G3DSA:3.40.366.10 (GENE3D).
Ø IPR016035: Acyl transferase/acyl hydrolase/lysophospholipase, represented by one signature: SSF52151 (SUPERFAMILY)
-
This is one domain with two catalytic active sites:
-
Active site residue (malonyl transferase; EC 2.3.1.39) = 581
- Active site residue (beta-hydroxyacyl dehydratase; possibly EC 4.2.1.61) = 878
Ø IPR016036: ACP-binding region of malonyl transacylase, represented by one signature: SSF55048 (SUPERFAMILY)
3)
Domain
Ø IPR013217: Methyltransferase type 12 domain, represented by one signature: PF08242 (PFAM).
4) Enoyl reductase domain (ER; EC 1.3.1.10):
Ø IPR011032: GroES-like domain, represented by one signature: SSF50129 (SUPERFAMILY).
-
The N-terminal domain of several alcohol dehydrogenases has a
GroES-like fold
Ø IPR013149: Alcohol dehydrogenase domain, represented by one signature: PF00107 (PFAM).
-
This is the enoyl reductase domain
-
Nucleotide binding site for NADPH = 1671-1688
Ø IPR016040: NAD(P)-binding domain, represented by two signatures: G3DSA:3.40.50.720 (GENE3D), SSF51735 (SUPERFAMILY)
-
This entry identifies domains that bind NAD or NADP. The enoyl
reductase domain is known to use NADPH.
5)
Beta-ketoacyl reductase domain (KR; EC 1.1.1.100):
Ø IPR002198: Represented by four signatures: PF00106 (PFAM), PR00080 (PRINTS), PS00061 (PROSITE), PTHR19410 (PANTHER).
-
Nucleotide binding site for NADPH = 1886-1901
Ø IPR016040: NAD(P)-binding domain, represented by two signatures: G3DSA:3.40.50.720 (GENE3D), SSF51735 (SUPERFAMILY)
-
This entry identifies domains that bind NAD or NADP. The
beta-ketoacyl reductase domain is known to use NADPH.
6)
Acyl carrier protein (ACP):
Ø IPR006162: Phosphopantheine attachment site, represented by one signature: PS00012 (PROSITE).
-
PROSITE predicts phosphopantheine attachment site for ACP to be
between residues 2151-2166 (known to be at 2156)
Ø IPR006163: Phosphopantheine-binding domain, represented by two signatures: PF00550 (PFAM), PS50075 (PROSITE).
Ø IPR009081: Acyl carrier protein-like domain, represented by two signatures: G3DSA:1.10.1200.10 (GENE3D), SSF47336 (SUPERFAMILY).
7)
Thioesterase domain (possibly EC 3.1.2.14):
Ø IPR001031: Thioesterase domain, represented by one signature: PF00975 (PFAM).
- Active site residues = 2308, 2481
The graphical match shows that human fatty acid synthase can be subdivided into nine regions (as covered by non-overlapping signatures in the table), which represent seven catalytic domains and the acyl carrier protein (ACP). The enzyme activities can be linked up with most of these regions (as given). In certain cases, active sites and binding sites have been identified by UniProt - this additional information is provided above.
In some cases, there are multiple entries covering the same sequence, but assigned to different entries. This is because, although the signatures cover the same sequence, some signatures hit additional proteins, which provides a family/sub-family grouping for these domains. For instance, IPR016035 covers the malonyl transacylase (MPT) domain (an acyl transferase), but the signature represents a structural domain with a 3-layer alpha/beta/alpha topology that is also found in acyl hydrolases and in lysophospholipases. Therefore it is the PARENT of the more specific entry IP001227, which only covers acyl transferases. IPR001227 is in turn the PARENT of the even more specific entry IPR014043 that only covers specific acyl transferases. This hierarchical scheme represents the evolutionary divergence of domain sequences.
Similarly, both IPR016038 and IPR016039 represent the thiolase-like family of domains, which includes the domain found in beta-ketoacyl synthase. This domain forms two subdomains, each represented by a different entry (IPR014030 and IPR014031).
Structural features
The remaining seven entries in
the table above give information on the known and predicted structure of this
protein. These entries presents known
structural data from the structural database PDB (green stripe) and the
structural classification databases CATH (pink stripe) and SCOP (black stripe) (the
names such as 1xktA02 are
derived from the PDB entry upon which they are based, here PDB entry 1xkt, chain A, region 2), as
well as predicted structural data from the homology model databases ModBase
(yellow stripe). The graphical match for
the PDB entries 2jfd, 2cq5, 1xkt display the full length of the original PDB entries, here
covering only small regions of the protein.
The ModBase homology model (MB_P49327) provides predicted structure for
the remainder for the protein.
The PDB entry 1xkt has been classified by both CATH and SCOP, although their
classification varies from one another.
SCOP (d1xkta) classifies the region covered by
1xkt as one functional domain having a 3-layer alpha/beta/alpha topology. CATH classifies
the region covered by 1xkt as two
structural subdomains, where 1xktA01
subdomain has a Rossmann-like fold and 1xktA02
insert subdomain has an orthogonal bundle fold.
What the Structure Tells Us
Structures associated with the human
fatty acid synthase can be viewed using AstexViewer®, which is linked from the
Match Table via the logo ![]() on the InterPro page (please note, there is no
link directly from this page to the AstexViewer®, therefore you need to go to
the
on the InterPro page (please note, there is no
link directly from this page to the AstexViewer®, therefore you need to go to
the ![]() link on the InterPro page for P49327).
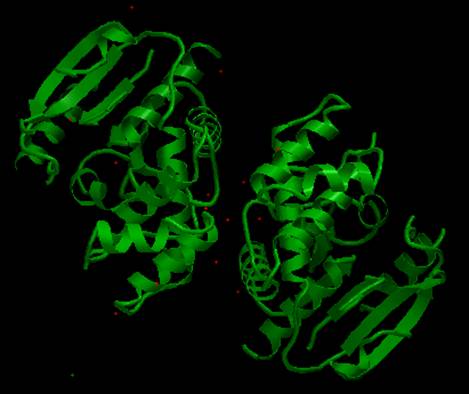
The AstexViewer® displays the full PDB structure for the thioesterase
domain. Note that there are two copies
of this domain present (i.e. shown as a dimer).
link on the InterPro page for P49327).
The AstexViewer® displays the full PDB structure for the thioesterase
domain. Note that there are two copies
of this domain present (i.e. shown as a dimer).
There are structures available for
several different fatty acid synthases from a variety of organisms in the
Protein Data Bank (PDB), both as type I multi-functional enzymes, as well as
type II individual enzymes. A detailed
description and visualisation of the structural features of fatty acid
synthases can be found at the PDB ‘Molecule of the Month’.
The crystallographic structures of various fatty acid synthases have
provided insight into their mechanism of action.
|
|
|
AstexView of human fatty acid synthase,
thioesterase domain: (shown as a dimer). |