Interpro 90.0 New Features And Bug Fixes
interproAnnouncement of the Pfam website decommission
After more than 20 years of service, we have decided to retire the Pfam website from October 2022. However, the Pfam data will continue to be available through the InterPro website, and the InterPro developers have been working hard over the last year to implement the vast majority of the Pfam website features into the InterPro website. More information can be found in this Pfam blog post.
New features
Exact search by gene identifier
As part of the InterPro text search feature, users can search for a protein accession (e.g. P05067). Following comments from one of our users, we have implemented the text search for a protein identifier (e.g. A4_HUMAN) which returns a link to the corresponding protein. Additionally, a gene name search (e.g. A4) retrieves the corresponding protein matches for key organisms.
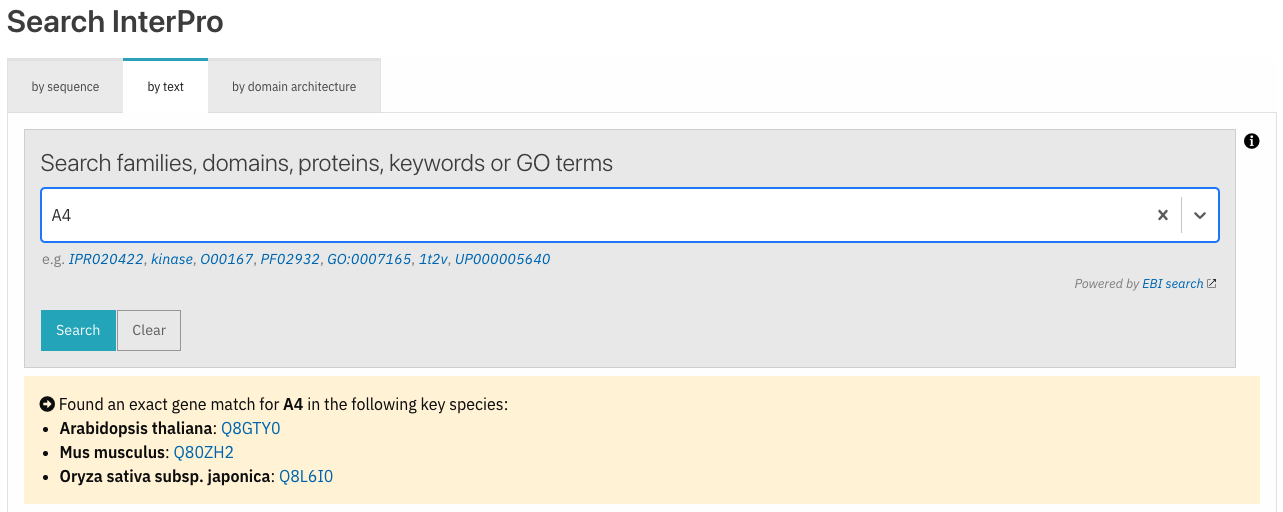 Figure 1. Exact match results for gene A4 search.
Figure 1. Exact match results for gene A4 search.
Downloads are saved
When using the Select and Download InterPro data feature, accessible through Results>Your downloads, the results were previously saved only for the current session, meaning that the user’s results were lost after closing the browser window. The results are now stored in the browser (IndexedDB), instead of just kept in memory, allowing the user to retrieve previous searches.
Re-running a sequence search
The InterPro sequence search is powered by InterProScan and the results are accessible under Results>Your InterProScan searches. By default, search results are kept in memory for 7 days, but they can be saved in the browser for retrieval after this period. When a search has been run using a previous version of InterProScan, the user can now re-run the search using the latest version.
New information on Pfam Set pages
Pfam clans (known as Set in InterPro) regroup evolutionary related signatures. On an InterPro Set page, different tabs allow access to a number of information. The Entries tab provides the list of entries included in the set (accession and name). For Pfam sets, we have added links to the signatures SEED alignment and domain architectures pages, as shown in Figure 2.
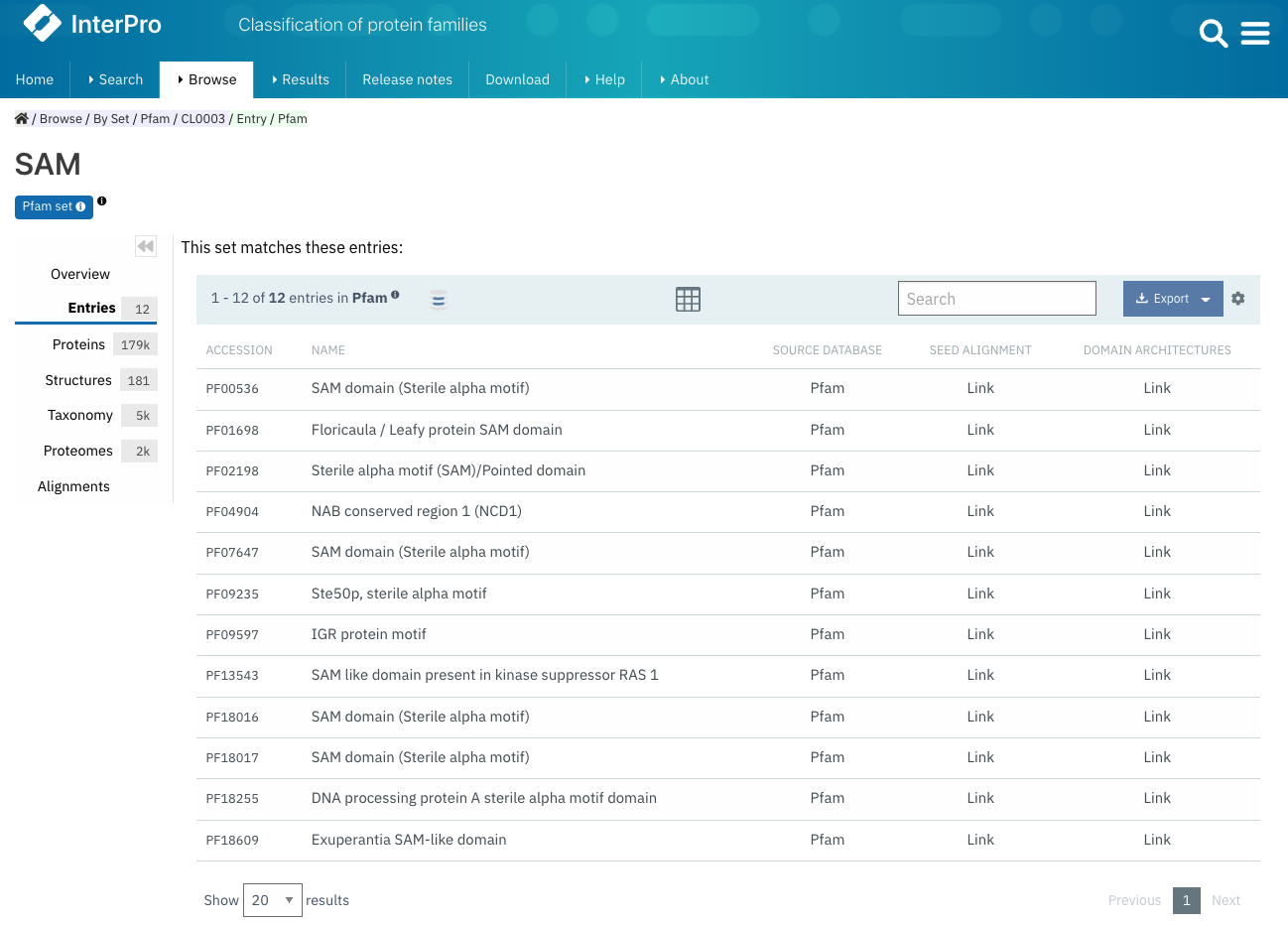 Figure 2. List of Pfam entries included in the Pfam SAM clan (CL0003) with links to the SEED alignment and domain architectures.
Figure 2. List of Pfam entries included in the Pfam SAM clan (CL0003) with links to the SEED alignment and domain architectures.
Reorganisation of the Download page
The Download page gives access to pre-calculated InterPro matches and the download of InterProScan. The page has been reorganised as tabs and now also includes Pfam files downloads, as shown in Figure 3.
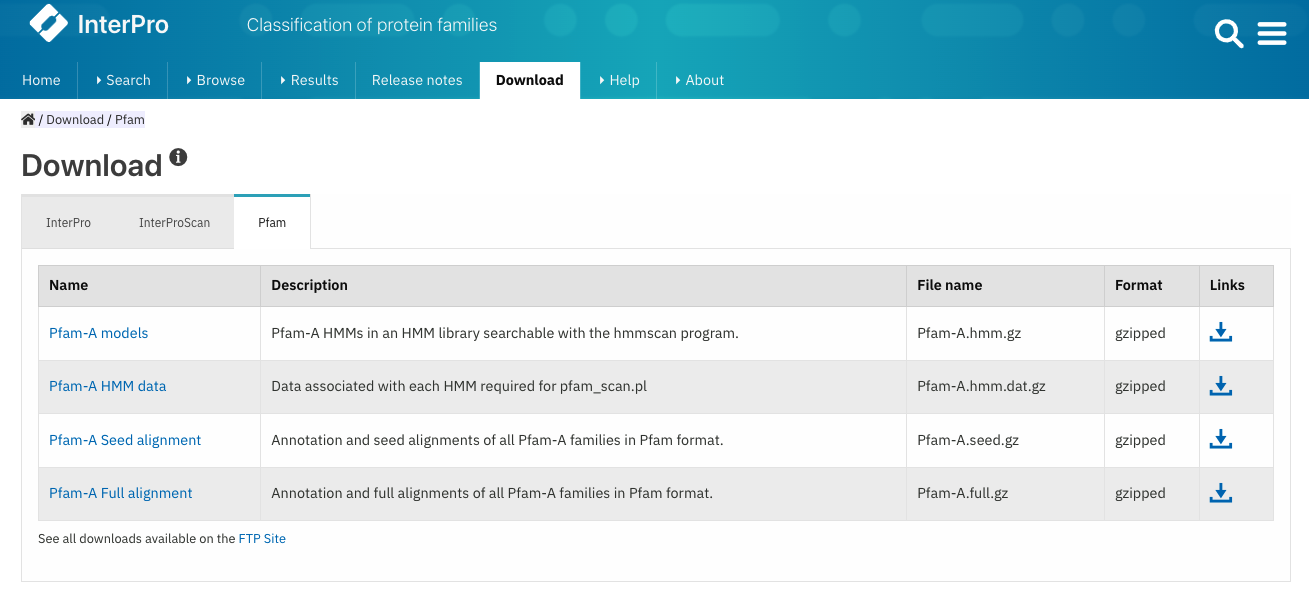 Figure 3. InterPro download page, Pfam tab.
Figure 3. InterPro download page, Pfam tab.
Addition of a documentation page for the settings
We have updated the InterPro documentation and added explanations regarding the parameters available from the InterPro website Settings page (accessible through the hamburger icon in the header).
Bug fixes
InterProScan vs protein viewer results differences resolved
Previously, the representation of InterProScan results for a sequence search for multi-domain proteins was different to the one shown in a protein page that had the exact same sequence. The difference was due to the way the locations for InterPro entries were calculated in the client, which differed from the way they were calculated in production. Basically, the production algorithm created two locations out of two non-overlapping signature matches, while the client was creating a single location that covered both. We have updated the client to behave the same way as the production algorithm.
Fix generation of FASTA format in Download
When downloading data in FASTA format using the generated code snippets, sequences with more than one hit were written several times because match locations were part of the header. Each sequence is now reported only once, whether an entry has multiple hits on the same protein, or multiple entries hit the same protein.
Other fixes
- Links for the download forms were not being escaped, this has been resolved.
- The signature logo and the taxonomy widget had some styling issues that have been corrected.
- Bugfix for the release notes links.
