Interpro 101.0: New Features And Improvements
interproThis release includes new features and improvements to the InterPro website. The list of changes is detailed in this post. If you have any feedback or suggestions, please contact us. For more regular updates, you can follow us on social media X and/or LinkedIn.
Post content
- Data updates
- Filtering InterPro entries in Browse
- Sorting lists of InterPro entries, member database signatures and clans/sets in Browse
- Signature matches for proteins and structures in clans/sets
- External links in Interactions subpage
- Upcoming changes to the sequence search
- Fixes
Data updates
In this release, we have updated NCBIfam to version 15.0 and updated InterPro entries accordingly. We also have integrated 753 new signatures from the NCBIfam (318), Pfam (446), PANTHER (2) and CDD (1) databases.
We have also updated Pfam-N data, available in the Other features section of the protein sequence viewer. This update uses Vision-Transformer Deep Learning Techniques, a different methodology compared to previous versions. The model has been trained using all InterPro member database signatures, including Pfam 37.0. More information about the model and its performance can be found in this Pfam blog post.
New features
Browsing InterPro data
In this update, we have made a few changes to enhance the user experience while browsing InterPro data from the Browse tab on the website menu.
Filtering InterPro entries
InterPro entries can now be filtered to only display the list of the latest entries that have been integrated in the current release version. This option is available from the option panel located on the left-hand side in Browse By InterPro (www.ebi.ac.uk/interpro/entry/InterPro/#table).
Sorting lists of InterPro entries, member database signatures and clans/sets
The list of entries available in Browse By InterPro, Browse by member DB and Browse by Clan/Set can be sorted by accession in ascending or descending order (Figure 1). This feature gives the possibility to display the latest entries first and avoid having to go through the whole list if the user is only interested in the entries added in the latest update of the database.
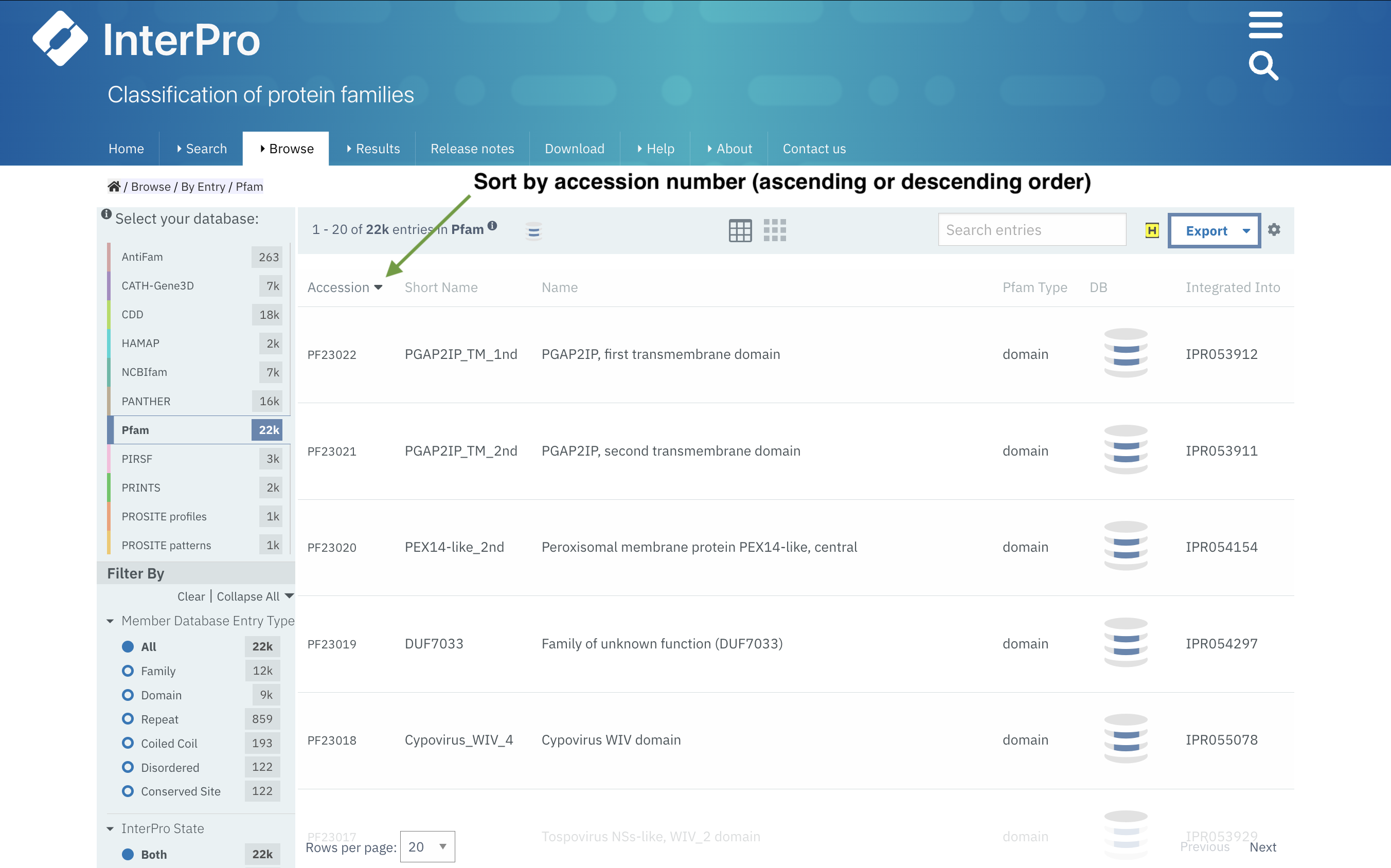 Figure 1. Example of Pfam signatures with accessions by descending order in Browse by member DB.
Figure 1. Example of Pfam signatures with accessions by descending order in Browse by member DB.
Signature matches in clans/sets
On the clan/set pages, the Proteins and Structures subpages feature tables displaying the proteins and structures that match the signatures included in the set/clan. The Proteins subpage table includes details such as the UniProt accession, name, species, gene, and AlphaFold structure prediction, along with the corresponding signature for each protein. Similarly, the Structures subpage table lists the PDB accession, name, image of the structure, and the specific signature that each structure matches (Figure 2).
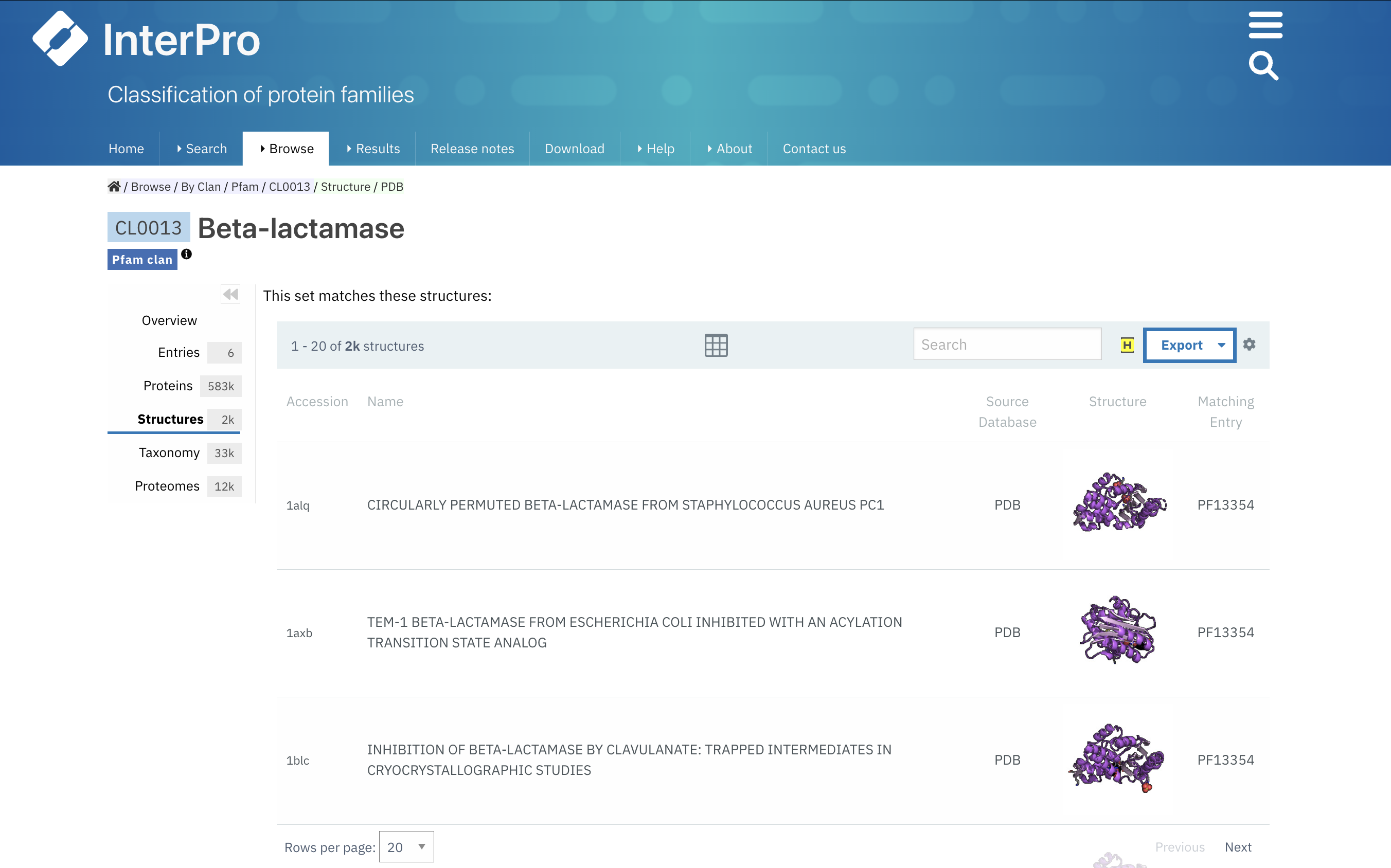 Figure 2. List of structures matched by the Pfam Beta-lactamase clan, with the matching Pfam signatures.
Figure 2. List of structures matched by the Pfam Beta-lactamase clan, with the matching Pfam signatures.
External links in Interactions subpage
Links to IntAct and EuropePMC for the IntAct entries listed in the Interactions subpage of InterPro entries have been added (example: IPR000719). The list of interactions can be sorted by Intact accession number, PMID, molecule A or molecule B. Additionally, each column can be filtered by name.
Upcoming changes to the sequence search
We are pleased to announce that on the next InterPro release (102.0), we will be supporting nucleotide sequence search through the InterPro sequence search on the website, alongside the reorganisation of jobs with multiple sequences. Currently, the nucleotide sequence search is only available through a local installation of InterProScan. Unfortunately, the new version will be incompatible with previously saved jobs in the browser. If you have any jobs you want to keep, please export them in JSON format, you will be able to import them back once the new version has been released.
Fixes
Links to Wikipedia articles in the infobox found in the Wikipedia section in Pfam entries are now handled.
The list of previous releases wasn’t in the correct order following release 100.0; this issue has been resolved.
The table counter indicator was not changing when changing pages in tables.
We fixed the query timeout error that was arising when browsing sets/clans.
The HMM logo settings are now displayed above the logo.
The shortcut to the Code snippet generator on the documentation page of the website (www.ebi.ac.uk/interpro/help/documentation) has been updated.
