 SRC, proto-oncogene tyrosine-protein kinase
SRC, proto-oncogene tyrosine-protein kinase
P12931 Human c-Src entry from InterPro:
|
Interpro Entry |
Method accession |
Graphical match |
Method name
|
|
|
Prot_kinase |
||
|
|
pkinase |
||
|
|
PROTEIN_KINASE_ATP |
||
|
|
PROTEIN_KINASE_DOM |
||
|
|
SH2 |
||
|
|
SH2 |
||
|
|
SH2DOMAIN |
||
|
|
SH2 |
||
|
|
SH2 |
||
|
|
TYRKINASE |
||
|
|
TyrKc
|
||
|
|
SH3 |
||
|
|
SH3 |
||
|
|
SH3DOMAIN |
||
|
|
SH3 |
||
|
|
SH3 |
||
|
|
PROTEIN_KINASE_TYR |
||
|
NONE: |
|
1a07A0 |
|
|
NONE: |
|
d1a07a_ |
|
|
NONE: |
|
d1kswa1 |
|
|
NONE: |
|
d1kswa3 |
What InterPro tells us
From the graphical match above, you can see that the signatures in InterPro are subdivided into groups. These groups give you information about the domain architecture of the protein, as well as its family relationships.
Starting with the domain architecture of Src, there are three major domains represented above: SH2, SH3 and the kinase catalytic domain. The last four entries in the table above are from the structural classification databases CATH and SCOP (the names such as d1kswa3 are derived from the PDB entry that they are based on; here PDB entry 1ksw, chain a). If you follow the links to these databases, you will find descriptions of the structural features and classification of these domains, as well as links to the corresponding PDB entries. There are also InterPro entries describing these domains. IPR000980 has five signatures representing the SH2 domain, as shown above. The SH2 domain can be found in many signal-transducing proteins besides the Src family, as well as in structural proteins such as cytoskeletal fodrin and tensin, and in small adaptor molecules such as Crk and Nck. IPR001452 has five signatures representing the SH3 domain. The SH3 domain can be found in a wide variety of intracellular and membrane-associated proteins, including proteins with enzymatic activity, non-catalytic adaptor proteins, and structural proteins. There are links to some of these proteins families within the section labelled ‘Found in’ in the IPR001452 entry. Each of the signatures making up IPR000980 and IPR001452 is representative of all the protein families found in these entries; the proteins from the Src family that contribute to these signatures are shown in the table. IPR008266 has one PROSITE signature representing the tyrosine protein kinase active site.
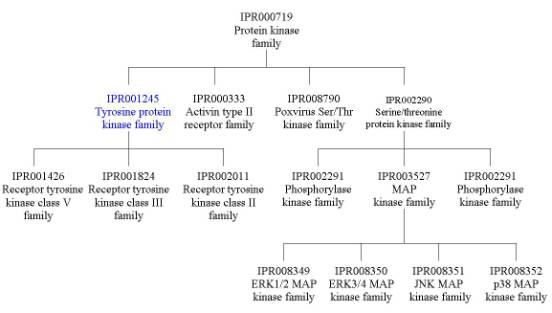
To look at the family relationships that involve Src, we need to start with the InterPro entry at the top of the table (IPR000719), which has four signatures representing the family of protein kinases. This is a very large family of proteins with 9,697 protein matches that represent the two types of protein kinases: serine/threonine-specific and tyrosine-specific (such as Src). These proteins share a number of conserved regions in the catalytic domain, which is why the signatures for IPR000719 overlap the Src catalytic domain (d1kswa3). If you follow the links to IPR000719, you will find that there are several InterPro families listed under the section labelled ‘Children’; this is because this large protein kinase family has been subdivided into smaller, more closely related families. To find out all the family relationships within the protein kinase family, you can either follow these links to the different InterPro entries, or you can follow the link labelled ‘[tree]’ found directly underneath the ‘Children’ tag (or follow the link provided here). One of the families listed under the ‘Children’ section is IPR001245, which represents the tyrosine protein kinase family containing Src. To give you an idea of the extent of the protein kinase family, look at the tree derived from IPR000719 below:
What the structure tells us
A description and visualisation of the structural features of Src can be found at the PDB database. The crystallographic structure of Src provided unparalleled insights into the mode of regulation of Src kinase activity.
Next: Table of Src family of proteins
Previous: How Src works