SV40
By Jennifer McDowall
|
|
|
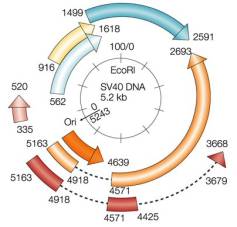
The SV40 Genome Reprinted by permission from Nature Reviews
Cancer 2:957-964, A. Gazdar, J. S. Butel and M. Carbone "SV40 and tumours: myth, association or
causality?" copyright (2002) Macmillan Magazines Ltd |
Polyomavirus are small, icosahedral, non-enveloped DNA viruses that infect a variety of vertebrate species. The name polyoma comes from the ability of mouse polyomavirus to induce multiple tumours in their infected host, these tumours arising from more than a dozen different cell types. However, not all polyomavirus induce tumours in their natural hosts under natural conditions. The various polyomavirus that have been studied appear to establish infections via the respiratory tract in newborn hosts, followed by chronic infection in the kidneys, with virus present in the urine. Three polyomavirus have been found in humans, Jamestown Canyon virus (JCV), BK virus (BKV) and simian virus 40 (SV40). JCV and BKV are both of human origin and can induce diseases under certain conditions: neurodegenerative leukoencephalopathy with JCV in AIDS patients, and tubulointerstitial nephritis with BKV in kidney transplant patients. Both JCV and BKV have possible roles in carcinogenesis. By contrast, SV40 is of monkey origin, but has been identified in specific human tumour cells. SV40 entered the human population as a contaminant of poliovirus vaccine in several countries between 1955 and 1963, because the vaccines were prepared in primary cultures of rhesus monkey kidney cells; subsequent vaccines are SV40-free. Monkey to human transmission may also spread SV40. SV40 is thought to be widely distributed in the human population.
Architecture of a virus
As with polyomavirus genomes, the SV40 genome consists of two sets of expressed genes: the early expressed genes, which encode proteins required for viral replication, including the small and large T antigens (small-t Ag, large-T Ag) and the 17KT protein; and the late expressed genes, which encode structural proteins necessary for viral assembly, including three coat proteins (VP1, VP2, VP3) and a maturation protein (agnoprotein). Different SV40 strains show variability within the variable domain of the T antigen proteins, as well as within the non-translated regulatory region that contains the origin of replication (ori) and promoters and enhancers of replication.
Alternative splicing produces two early proteins, large-T Ag and small-t Ag. The large-T Ag functions as a replicative helicase, binding to specific sites in the ori to promote the local unwinding of DNA, as well as recruiting cellular DNA replication proteins to the site, including topoisomerase I, replication protein A, and DNA polymerases. The large-T Ag autoregulates the early promoter, and can indirectly contribute to the activation of late transcription. The large-T Ag also plays a role in viral maturation by influencing the phosphorylation of capsid proteins. The small-t Ag plays a key role in infections by increasing the level of virus during the lytic cycle, and by enhancing cell transformation by the large-T Ag.
The DNA-binding maturation protein, agnoprotein, is thought to be involved in viral assembly or release, and acts to increase the efficiency of plaque formation. The agnoprotein from JCV, which shows 50-60% homology to SV40 agnoprotein, co-localizes with cytoskeletal tubulin, suggesting a role in the stabilization of microtubules during infection.
The viral capsid is composed of pentamers of VP1. Two minor structural proteins, VP2 and VP3, produced by alternative splicing, bridge VP1 capsid to the SV40 genome. VP3 is essential for formation of infection particles, and may be involved in virus-cell interactions during post-packaging steps.
Next: SV40 and cancer