DNA Ligase IV
By
Jennifer McDowall
To view structure of DNA ligase
Double-strand breaks (DSBs) in DNA pose a lethal threat to a cell: if not repaired, DSBs can cause cell death; if repaired incorrectly, DSBs can cause mutations and chromosomal rearrangements that can lead to the loss of growth control and cancer. DSBs occur naturally as a result of oxidative metabolism and V(D)J recombination in lymphocytes, as well as in response to ionising radiation and to chemotherapeutic agents such as bleomycin.
Cells have evolved a number of ways of dealing with the damage arising from DSBs: homologous recombination (HR), non-homologous end joining (NHEJ), and single-strand annealing. In mammalian cells, HR and NHEJ predominate. HR is the major repair pathway following DNA replication, using an undamaged sister chromatid as a template to accurately repair the damage. NHEJ is used to repair DSBs arising from DNA-damaging agents such as ionising radiation, and for those arising from V(D)J recombination. Unlike HR, NHEJ is an inherently error-prone process, whereby two DNA ends are joined directly without the need for sequence homology, although in some cases small regions of homology may exist in the single-stranded overhangs in the break. NHEJ can result in the loss of nucleotides from the site of breakage, which can be deleterious if the break occurs in a gene or its controlling sequences; yet this may still prove more beneficial to a cell than leaving the breaks unrepaired. There are several proteins known to be involved in NHEJ, one of which is DNA ligase IV.
|
|
|
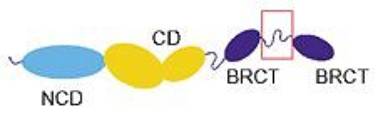
Domain structure of DNA ligase IV. The structure of the red-boxed region has been solved by crystallography. Reprinted from Genome Biology 3(4), I. V. Martin and S. A. MacNeill, ATP-dependent DNA ligases, 2002, REVIEWS3005, PMID: 11983065. |
DNA Ligase IV, a Repair Protein
DNA ligase IV, which is conserved in all eukaryotes, is part of a family of ATP-dependent DNA ligases that are involved in DNA replication, recombination and repair. These DNA ligases have two common domains: a catalytic domain (CD) that contains several conserved nucleotide-binding motifs, and a conserved non-catalytic domain (NCD). In addition, DNA ligase IV has a long C-terminal extension comprising of two BRCT domains (after the C-terminal domain of a breast cancer susceptibility protein, BRCA1), which are phosphopeptide-binding modules found in many proteins that regulate DNA damage responses, such as BRCA1, MDC1 and BARD1. A short linker region that is required for the binding of the XRCC4 protein, which is important for ligase activity, connects the two BRCT domains.
DNA ligase IV is a nuclear enzyme that joins the breaks in the phosphodiester backbone of DNA. The reaction mechanism involves the formation of a covalent enzyme-AMP intermediate from the cleavage of ATP to AMP and pyrophosphate. The adenylate group from AMP is then transferred to the 5’-phosphate of the nicked DNA molecule. Finally, the DNA ligase seals the gap by phosphodiester bond formation, via the displacement of the AMP residue with the 3’-hydroxyl group from the adjacent DNA strand.
Next: NHEJ, How It Works