Sm Ribonucleoproteins
By
Jennifer McDowall
Messenger RNA (mRNA) acts as the vital link between the genetic information stored in DNA and the protein products required throughout an organism. In eukaryotes, the progress of mRNA from the time of its synthesis as it is transcribed on the DNA genome, to its translation into protein products in the cytoplasm, can involve several processing steps. When mRNA is first transcribed as pre-mRNA, it contains both exons (translated sequences) and introns (intervening sequences). The presence of introns provides an important mechanism for flexibility in protein sequences, as a series of different proteins specialised in function or tissue-specificity can be produced from one gene by alternative splicing, whereby introns as well as some exons can be spliced out differently from the various copies of a transcript. Consequently, one of the major post-transcriptional processing events involves splicing the introns out of pre-mRNA, the discovery of this process earning a Nobel Prize in Physiology and Medicine for Philip Sharp and Richard Roberts in 1993.
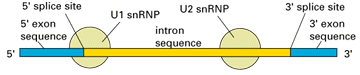
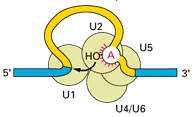
Specialised spliceosomes splice the introns out of pre-mRNA and seal the exon ends together, using the splicing consensus sequences at the intron/exon boundaries to identify the correct positions to splice. Sometimes a regulatory protein will mask a splicing sequence, resulting in alternative splicing. The spliceosomes consist primarily of RNA-protein complexes called small nuclear ribonucleoproteins (snRNPs). The snRNPs are composed of small nuclear RNAs (snRNAs) - U1, U2, U4, U5 and U6 - as well as a group of seven proteins known as Sm ribonucleoproteins that collectively make up the extremely stable Sm core of the snRNP. The snRNPs bind to the pre-mRNA in a specific order to align the splice sites for cleavage, which involves RNA-RNA pairing between the snRNA and the pre-mRNA with the help of the Sm proteins. The U1 snRNP binds to the 5’ end of the intron and the U2 snRNP binds close to the 3’ end of the intron (at the branch point), followed by the binding of the U4/U6 snRNPs that play an important role close to the reaction centre, and finally the U5 snRNP that helps hold the two exons together. After the intron is spliced out it is rapidly degraded, and the two exons are ligated together.
|
|
|
|
U1 and U2 snRNPs bind first
to intron ends on pre-mRNA |
Other snRNPs bind, cleave and rejoin mRNA |
Sm
Ribonucleoproteins, RNA Processors
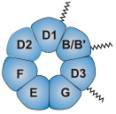
The Sm core of U1, U2, U4, and U5 snRNPs consists of seven proteins: B/B’ (alternative splicing products), D1, D2, D3, E, F and G. These proteins gained their name from their reaction with Sm serotype antibodies from patients with the autoimmune disease systemic lupus erythematosus. The Sm proteins form a seven-member ring core structure that encircles the RNA. All the Sm proteins share a conserved Sm motif, consisting of two sets of conserved sequences (Sm1 and Sm2) separated by a large loop region, which appear to be the sites of protein-protein interactions necessary for the core structure to form. The Sm core is essential for the biogenesis, transport and function of the snRNP particles.
|
|
7-member ring Sm core Reprinted from Cellular and Molecular Life Sciences (Birkhäuser Verlag, Basel, Switzerland) Oct 61(19-20), D. Schumperli and R. S. Pillai, The Special Sm Core Structure of the U7 snRNP: Far-reaching Significance of a Small Nulcear Ribonucleoprotein, pp2560-1570, 2004, PMID: 15526162. |
Next: Sm Core Assembly and
Specificity