Sm Ribonucleoproteins
Sm Core Assembly and Specificity
The assembly of the Sm core occurs as an ordered, protein-assisted process in the cytoplasm after the nuclear export of nascent snRNAs. The proper assembly of the Sm core, as well as the 5’-cap hypermethylation and 3’-end processing of the snRNAs are required before the snRNPs can be transported back into the nucleus to function in pre-mRNA splicing.
The Survival of Motor Neurons (SMN) protein plays an important role in regulating the assembly and specificity of Sm proteins, and appears to be sufficient for Sm core assembly. The crucial role that SMN plays is illustrated by cases where it is mutated, resulting in the fatal neuromuscular disorder, spinal muscular atrophy (SMA), characterised by the degeneration of spinal cord motor neurons. SMN is a multi-protein complex that includes several proteins, such as Gemins2-7, where Gemin3 acts as an RNA helicase. The SMN complex acts as a scaffold on which the Sm and snRNA components are assembled in a sequence-specific manner. Some Sm proteins (B, D1, D3) bind directly to SMN through their RG (arginine- and glycine-rich) domains, while snRNAs contain both SMN-binding domains as well as U-rich Sm-binding sites. SMN acts as a specificity chaperone to ensure the Sm core assembles onto the correct snRNA, by only incorporating SMN-bound Sm proteins and sequence-specific snRNAs into snRNPs. This stringent control prevents the deleterious association of Sm proteins with other RNAs, which could interfere with their functioning. In this way the SMN complex determines the types of snRNPs assembled.
Lsm Proteins, a Family with
Diverse Capabilities
The have been several proteins identified in many organisms, including bacteria, that are related to Sm proteins in sequence, structure and function, which together form the Lsm (Like-Sm) protein family. Lsm proteins contain Sm motifs; form 6- or 7-member rings with a central hole through the RNA may pass; and are involved in various essential RNA-processing tasks. For instance, bacterial Hfq proteins facilitate RNA-RNA interactions, archaeal Lsm proteins can associate with RNase-P RNA required during ribosome synthesis and function, and eukaryotic Lsm proteins are involved in pre-mRNA splicing, histone 3’-end formation, tRNA processing, rRNA maturation, mRNA degradation and telomeric DNA synthesis.
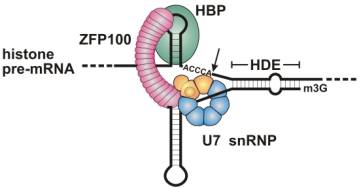
In eukaryotes, different snRNPs can contain unique sets of Lsm proteins, because the Sm-binding sites on snRNAs are not equivalent. The different combinations of Lsm proteins allow a cell to create different complexes with distinct functions involved in a variety of cellular events. For instance, U7 snRNPs contain two unique Lsm proteins, Lsm10 and Lsm11 (replace D1 and D2), which are required for histone RNA processing. Lsm1-7 form a cytoplasmic rather than a nuclear complex that mediates RNA decapping and degradation. Lsm2-7 associates with snR5 snoRNA required for the site-specific pseudouridylation of rRNA. These diverse functions show the importance of Lsm proteins as modulators of RNA biogenesis and function.
|
|
Molecular
mechanism of histone pre-mRNA 3’ end processing by U7 snRNP. Reprinted from Cellular and Molecular Life Sciences (Birkhäuser Verlag, Basel, Switzerland) Oct 61(19-20), D. Schumperli and R. S. Pillai, The Special Sm Core Structure of the U7 snRNP: Far-reaching Significance of a Small Nulcear Ribonucleoprotein, pp2560-1570, 2004, PMID: 15526162. |
Next: What InterPro Tells Us
Previous: Sm Ribonucleoproteins,
RNA Processors