TRK Receptors
Trk Receptor Composition
The Trk receptors share a fairly similar composition. The extracellular region consists of a leucine-rich repeat flanked by two cysteine-rich domains involved in protein-protein interactions, as well as an immunoglobulin-like domain involved in neurotrophin ligand binding. These extracellular domains not only bind ligand, but are also involved in Trk receptor dimerisation. The intracellular region consists of a tyrosine protein kinase domain plus several tyrosine-containing motifs similar to those found in other receptor tyrosine kinases.
Trk receptors dimerise in response to ligand binding and phosphorylated one another at tyrosine residues, which in turn increases the activity of the kinase domain and provide attachment sites for signalling molecules, which themselves become phosphorylated by the receptor tyrosine kinase. A number of different pathways can ultimately be activated through the recruitment of different signalling molecules, and can interact with many additional molecules implicated in neuronal survival, morphological development, or differentiation.
What InterPro Tells Us
Q16288 Human TrkC Receptor
InterPro Domain Architecture:
![]()
|
InterPro Entry |
Method Accession |
Graphical Match |
Method Name |
|
IPR000372 |
PF01462
|
|
LRRNT |
|
IPR000372 |
SM00013
|
|
LRRNT |
|
IPR000483 |
SM00082 |
|
LRRCT |
|
IPR000719 |
PD000001
|
|
Prot_kinase |
|
IPR000719 |
PS00107 |
|
PROTEIN_KINASE_ATP |
|
IPR000719 |
PS50011 |
|
PROTEIN_KINASE_DOM |
|
IPR001245 |
PR00109 |
|
TYRKINASE |
|
IPR001245 |
SM00219 |
|
TyrKc |
|
IPR001611 |
PF00560 |
|
LRR_1 |
|
IPR001611 |
PR00019 |
|
LEURICHRPT |
|
IPR002011 |
PS00239 |
|
RECEPTOR_TYR_KIN_II |
|
IPR003599 |
SM00409
|
|
IG |
|
IPR007110 |
PF00047 |
|
ig |
|
IPR007110 |
PS50835 |
|
IG_LIKE |
|
IPR008266 |
PS00109 |
|
PROTEIN_KINASE_TYR |
|
IPR011009 |
SSF56112 |
|
Kinase_like |
|
Classification |
PDB Chain/ Domain ID |
PDB Chain/Structural Domains |
|
|
1wwc |
1wwca |
|
|
|
2.60.40.10.38 |
1wwcA0 |
|
|
|
b.1.1.4 |
d1wwca_ |
|
|
From the graphical match above, you can see that the signatures (method accession) are divided into ten InterPro entries for human TrkC. These signatures give information on the family relationships and domain architecture of this protein.
The InterPro entry IPR002011 has one signature representing the receptor tyrosine protein kinase class II family: PS00239 from the PROSITE database, which based its signature on the putative site for auto-phosphorylation (including the tyrosine residue thought to be auto-phosphorylated) within the kinase domain, which appears to be highly conserved among class II tyrosine protein kinases. The InterPro entry gives details regarding the taxonomic distribution of different tyrosine protein kinase class II proteins.
Eight of the InterPro entries above give information
on the domain architecture of human TrkC.
IPR000372 has two signatures representing the
cysteine-rich domain that flanks the N-terminal of the leucine-rich repeat
(LRR) region: PF01462 from the PFAM database, and SM00013 from the SMART database. The next domain is represented by IPR001611,
which covers the leucine-rich repeat (LRR)
region, here showing two motifs in the repeat.
IPR001611 has two signatures: PF00560 from the PFAM database, and PR00019 from the PRINTS database. IPR000483 represents the second cysteine-rich domain that flanks the C-terminal
of the LRR region, and which is represented here by one signature: SM00082 from the SMART database. Following the LRR and its flanking regions
comes the immunoglobulin-like domain.
This domain is represented by two InterPro entries: IPR003599 with one signature (SM00409 from the SMART database) that represents a domain related to an
immunoglobulin subtype, and IPR007110 with two signatures (PF00047 from the PFAM database and PS50835 from the PROSITE database) that represents immunoglobulin-like
domains. IPR007110 is a parent of
IPR003599, as it covers the same sequence, but includes a broader category of
immunoglobulin-like domains (parent/child relationships can be viewed in the
InterPro entries for these domains, providing a link between related
signatures). The next domain found is
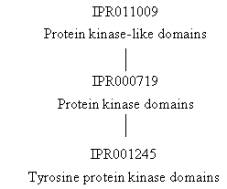
the protein kinase domain, which is represented by three InterPro entries: IPR011009 with one signature (SSF56112 from the Superfamily database) that represents the protein kinase-like
domain, IPR000719 with three signatures (PD000001 from the ProDom database, and PS00107 and PS50011 from the PROSITE database) that
represent the protein kinase domain, and IPR001245 with two signatures (PR00109 from the PRINTS database and SM00219 from the SMART database) that represent the tyrosine protein
kinase domain. All three of these
entries cover the same sequence, with IPR011009 being the broadest category
representing domains with a similar protein kinase structure (even if they
diverge in sequence), IPR000719 representing protein kinase domains related in
sequence, and IPR001245 representing specifically tyrosine-type protein kinase
domains. Their relationships are IPR011009
parent of IPR000719 parent of IPR001245:
|
|
The
final InterPro entry, IPR008266, represents the tyrosine protein kinase
active site, which has one signature: PS00109 from the PROSITE database, which centers
around a conserved aspartic acid residue known to be important for the
enzymatic catalytic activity of tyrosine protein kinases.
The remaining three entries in the table above are from the structural database PDB (green stripe), and from the structural classification databases CATH (pink stripe) and SCOP (black stripe) (the names such as d1wwca_ being derived from the PDB entry upon which they are based, here PDB entry 1wwc, chain A). The graphical match for the PDB entry 1wwc displays the length of the original PDB entry. The region covered by the PDB is the ligand-binding region responsible for binding to neurotrophin (which is neurotrophin-3 in the case of human TrkC). There are no signatures covering this region, possibly because it varies between different types of TRK receptors to give the proteins their ligand specificity. The CATH (1wwcA0) and SCOP (d1wwca_) databases give information on the classification of this region.
What the Structure Tells Us
Structures
of the ligand-binding domain of several TRK receptors (TrkA, TrkB and TrkC)
from a variety of species have been determined. Structures for individual proteins can be viewed using
AstexViewer®, which is linked from the InterPro Match Table of each protein via
the logo ![]() (please
note, there is no link directly from this page to the AstexViewer® for the
protein discussed above, therefore you need to go to the link on the InterPro
page for Q16288). The AstexViewer® displays the PDB structure
with the particular CATH or SCOP domain highlighted in yellow. There are many structures associated with
the ligand-binding domains of TRK receptors in the Protein Data Bank
(PDB). A detailed description and
visualisation of the structural features of TRK receptors can be found at the PDB ‘Molecule of the Month’,
providing insights into the molecular basis of action for these receptors.
(please
note, there is no link directly from this page to the AstexViewer® for the
protein discussed above, therefore you need to go to the link on the InterPro
page for Q16288). The AstexViewer® displays the PDB structure
with the particular CATH or SCOP domain highlighted in yellow. There are many structures associated with
the ligand-binding domains of TRK receptors in the Protein Data Bank
(PDB). A detailed description and
visualisation of the structural features of TRK receptors can be found at the PDB ‘Molecule of the Month’,
providing insights into the molecular basis of action for these receptors.