Cytochrome P450
Family Ties
Cytochrome P450 proteins were initially grouped into two general classes:
· Class I (or E) enzymes are primarily eukaryotic microsomal enzymes (2-component system). These enzymes were found to be integral membrane proteins that interact with NADPH-dependent FAD/FMN-containing reductases (CPR). Class E enzymes can be further subdivided into five sequence clusters, I-V.
· Class II (or B) enzymes are bacterial/mitochondrial/fungalCYP55. These enzymes were found to be freely soluble proteins that interact with NADH-specific FAD-containing reductases (3-component system).
However, the diversity of CYP proteins appears to be more complex than originally thought, some falling outside of this basic two-class system. There have been a few thousand cytochrome P450 enzymes identified. The current nomenclature system uses CYP, followed by a number (family), then a letter (subfamily), and another number (protein); e.g. CYP3A4. Each subfamily represents a cluster of tightly linked genes, these clusters being spread throughout the genome. In general, family members should share >40% identity, while subfamily members should share >55% identity. To date, there have been 58 CYP genes identified in humans (plus several pseudogenes), which fall into 18 families (40 subfamilies).
What InterPro Tells Us
P08684 Human cytochrome P450 CYP3A4
InterPro Domain Architecture
![]()
InterPro Entry |
Signatures |
Graphical Match |
Method Name |
|
IPR001128 |
G3DSA:1.10.630.10 |
|
GTP_EFTU |
|
IPR001128 |
PF00067 |
|
ELONGATNFCT |
|
IPR001128 |
PR00385 |
|
EFACTOR_GTP |
|
IPR001128 |
PS00086 |
|
GTP_EFTU_D3 |
|
IPR001128 |
SSF48264 |
|
GTP_EFTU_D2 |
|
IPR002401 |
PR00463 |
|
EF-1_alpha |
|
IPR008072 |
PR01689 |
|
Translat_factor |
|
Structural Features |
|
|
|
|
1tqn |
1tqnA |
|
|
|
a.104.1.1 |
d1tqna_ |
|
|
From the graphical match above, you can see that the signatures are all grouped into three InterPro entries for human cytochrome P450 CYP3A4. InterPro entries group together all the signatures that represent the same sequence found in the same set of proteins. These entries provide a family hierarchy of cytochrome P450 proteins.
FAMILY Entries
Ø IPR001128: Cytochrome P450 family, represented by five signatures: G3DSA:1.10.630.10 (GENE3D), PF00067 (PFAM), PR00385 (PRINTS), PS00086 (PROSITE), and SSF48264 (SUPERFAMILY).
Ø IPR002401: E-class P450 group I family, represented by one signature: PR00463 (PRINTS).
Ø IPR008072: E-class P450 CYP3A family, represented by one signature: PR01689 (PRINTS).
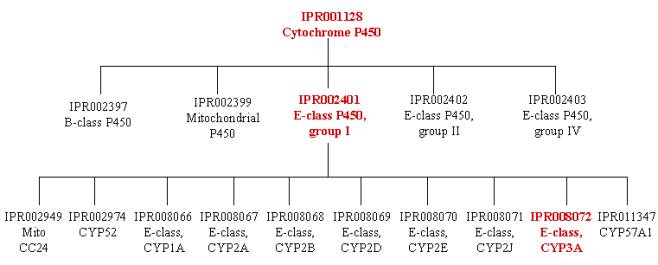
The three family groupings are hierarchical, covering the same sequence in expanding sets of proteins. The first entry, IPR001128, is top of the hierarchy representing the entire cytochrome P450 superfamily. As such, its signatures hit the largest group of proteins. This entry has five ‘children’ entries that cover the same sequence, but in different non-overlapping sets of proteins. These children are all shown in the tree below; the one that relates to the above protein is IPR002401, E-class P450, group I enzyme family. IPR002401 can be further subdivided, and itself has ten ‘children’ entries shown in the tree below; the one that relates to the above protein is IPR008072, E-class P450, CYP3A subfamily.
The remaining two entries in the table above give information on the structure of this protein, presenting known structural data from the structural database PDB (green stripe) and the structural classification database SCOP (black stripe) (the names such as d1tqna_ are derived from the PDB entry upon which they are based, here PDB entry 1tqn, chain A). The graphical match for the PDB entry 1tqn displays the full length of the original PDB entry, here covering the entire protein. The SCOP entry breaks down the PDB data into its structural domains (here just one domain), providing a structural classification for the protein domain and relating it to other structures in the PDB database. SCOP describes this domain as the cytochrome P450 domain, which has an all-alpha structure that is multi-helical in nature.
What the Structure Tells Us
Structures
associated with various cytochrome P450 can be viewed using AstexViewer®, which
is linked from the Match Table via the logo ![]() on the InterPro page (please note, there is
no link directly from this page to the AstexViewer®, therefore you need to go
to the
on the InterPro page (please note, there is
no link directly from this page to the AstexViewer®, therefore you need to go
to the ![]() link on the InterPro page for P08684). The AstexViewer® displays the PDB structure with the SCOP domain
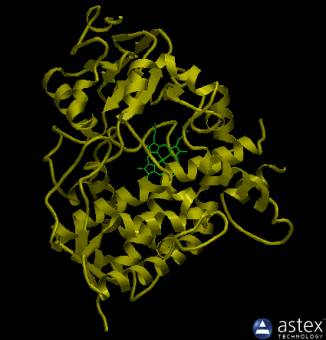
highlighted in yellow, as shown below.
Note, the haem iron in the core of the molecule is displayed in green.
link on the InterPro page for P08684). The AstexViewer® displays the PDB structure with the SCOP domain
highlighted in yellow, as shown below.
Note, the haem iron in the core of the molecule is displayed in green.
|
|
|
AstexView of human CYP3A4: the multi-helical protein contains a haem at its core (green). |
There are
structures available for various cytochrome P450 proteins from different
organisms in the Protein Data Bank (PDB).
A detailed description and visualisation of the structural features of
these proteins can be found at the PDB ‘Molecule of the Month’.
The crystallographic structures of various cytochrome P450 proteins have
provided insight into their mode of action.
Next: Table of Cytochrome P450 Enzymes in Humans
Previous: Cytochrome P450
and Cancer