AAA ATPases
Protein
Digestion in the Proteasome
|
|
|
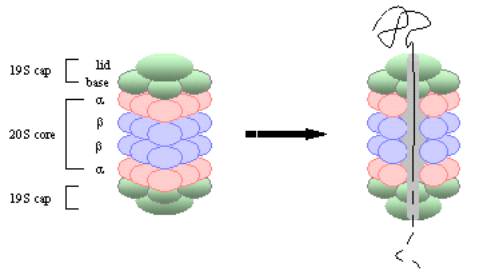
Left: The 30S proteasome consists of a 20S core and two 19S regulatory caps. Right: An ubiquitin-tagged protein is unfolded and de-ubiquitinated in the cap, then threaded through the core, where it is digested into peptides. |
In eukaryotes, proteasomes are large, ATP-dependent complexes located in both nucleus and cytosol. The 26S proteasome is composed of a 20S core that is responsible for degrading proteins, flanked at one end by a regulatory cap (the 30S proteasome has two 19S caps, one at either end). The 20S core is a hollow cylinder consisting of two outer a-rings and two inner b-rings, each ring type composed of seven distinct, homologous protein subunits (see figure above). Protease degradation is carried out by three of the b-ring proteins, their proteolytic sites facing the hollow interior cavity through which the condemned protein travels. Each b-ring protease is a threonine protease with distinct substrate specificities (one chymotrypsin-like activity, one trypsin-like activity and one post-glytamyl activity), to degrade as many types of proteins as possible. To protect the cell, these proteases are synthesized as inactive zymogens, which are cleaved to produce active enzymes once safely housed within the proteasome complex. The proteases breakdown condemned proteins into small peptides of 3-25 amino acids each.
Archaeal cells possess simpler ATP-dependent 20S proteasomes, consisting of just two proteins, a and b, with 14 copies of each arranged in a similar ring formation. Bacterial cells do not contain 20S proteasomes, but they do have large, compartmentalised protease complexes associated with AAA ATPases that perform the same function.
The Regulatory
Cap
Proteins to be degraded need to enter the small channel in the 20S core formed by the a-subunits. However, the entrance is usually blocked by the N-termini of the a-subunits, which together form a gate to control entry and prevent indiscriminate protein degradation. When a regulatory cap interacts with the gated end of a 20S proteosome core, it causes a conformational change in the proteosome that opens a passageway into the hollow centre of the complex.
Even when the gate into the core proteasome is open, the channel is very narrow, admitting only unfolded, linear proteins. It is therefore the job of the regulatory cap to unfold the condemned protein and to remove the ubiquitin chain, before it threads it through the core proteasome. The 19S regulatory cap can accomplish this; therefore, any protein to be degraded must first associate with this regulatory cap.
The 19S cap consists of 20 protein subunits, which when combined with the 20S core makes a 26S proteosome (or a 30S proteasome if there is a 19s cap at either end). The 20 subunits in the cap are arranged into two subcomplexes: the lid and the base. The lid consists of a ring of eight proteins, which disassembles the ubiquitin chains for re-use. The base contains a ring of six homologous AAA ATPases, which act like chaperones to carry out ATP-dependent unfolding of tagged proteins prior to being threaded into the proteosome core. Each AAA ATPase performs a distinct function within the complex, aiding in the recognition and degradation of different types of proteins. In addition, each step, from de-ubiquitination, and unfolding to threading the condemned protein into the 20S core, is regulated in some way by the ATPase complex.
Next: What InterPro Tells Us
Previous: AAA
ATPases, Driving Selective Protein Degradation