AAA ATPases
Family Ties
AAA ATPases are found in all living organisms and in all cellular compartments. They are a functionally diverse group of proteins within the P-loop-type NTPases, participating in a variety of cellular processes, including protein degradation, protein folding, protein translocation, mitosis, DNA replication and repair, and membrane fusion. AAA ATPases contain one or two conserved ATP-binding domains, which contains two conserved motifs, Walker A and Walker B. These ATP-binding domains are often attached to various other functional domains. Most AAA ATPases assemble into oligomeric (usually hexameric) ring-shaped structures with a central pore, and are usually involved in protein folding/unfolding and assembly/disassembly of protein complexes through ATP-dependent conformational changes. The functional variety seen between AAA ATPases is in part due to their extensive number of accessory domains and factors, and to their variable organisation within oligomeric assemblies, in addition to changes in key functional residues within the ATPase domain itself.
AAA ATPases are one member of the larger AAA+ ATPase superfamily. Arabidopsis thaliana show the greatest diversity of AAA+ ATPases, producing over 140 different AAA+ ATPase proteins. Most eukaryotes produce 50-80 different AAA+ ATPase proteins. AAA+ ATPases have been divided into a number of clades for classification, primarily based on structural elements in the proteins. AAA ATPases involved with protein degradation in proteasomes are part of clade 3. Clade 3 proteins usually form closed hexameric assemblies, and are involved in membrane fusion, microtubule severing, peroxisome biogenesis, and protein degradation. All these proteins make use of a molecular motor, coupling ATP-binding and hydrolysis to changes in conformational states in order to act upon a target substrate. The diversity within this clade is most likely due to determinants outside the ATPase core.
What InterPro Tells Us
P62191 Human AAA ATPase, 26S Protease Regulatory
Subunit 4
InterPro Domain Architecture
![]()
InterPro Entry |
Signatures |
Graphical Match |
Method Name |
|
IPR003593 |
SM00382 |
|
AAA |
|
IPR003959 |
PF00004 |
|
AAA |
|
IPR003960 |
PS00674 |
|
AAA |
|
IPR005937 |
TIGR01242 |
|
26Sp45 |
|
Structural Predictions |
|
|
|
|
MB_P62333 |
|
|
|
From the graphical match above, you can see that the signatures are all grouped into four InterPro entries for human AAA ATPase, 26S protease regulatory subunit 4. These entries provide a family grouping of this ATPase, as well as demarcating the ATPase core domain.
FAMILY Entries
Ø IPR005937: 26S proteasome subunit P45 family, represented by one signature: TIGR01242 (TIGRFAM).
DOMAIN Entries
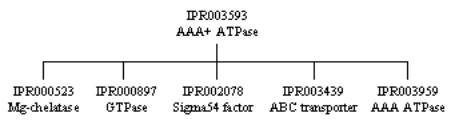
Ø IPR003593: AAA+ ATPase core domain (which is found in several other protein families as well), represented by one signature: SM00382 (SMART).
Ø IPR003959: AAA ATPase core domain, represented by one signature: PF00004 (PFAM).
Ø IPR003960: AAA ATPase active site subdomain, represented by one signature: PS00674 (PROSITE).
The three domain entries are related to one another, as they cover the same sequence but hit overlapping sets of proteins, and therefore form a hierarchical relationship. The first entry, IPR003593, is top of the hierarchy as it hits the most proteins, matching the AAA ATPase core domain found in AAA+ ATPase superfamily of P-loop-type NTP-binding proteins. It has five ‘children’ entries that cover the same sequence, but in different non-overlapping sets of proteins. These children are all shown in the tree below; the one that relates to the above entry is IPR003959, which describes the AAA ATPase core domain found only in those proteins that form the AAA ATPase family. Finally, IPR005937 represents a conserved region of 220 amino acids that contains the ATP-binding site in AAA ATPases.
The remaining entry in the table above provides a prediction
of the structure of this AAA ATPase, MB_P62333, based on a homology model from
the database ModBase (yellow stripe). There
is currently no PDB structure for this particular protein.
What the Structure Tells Us
Structures
for bacterial AAA+ proteases are available in the Protein Data Bank (PDB). A detailed description and visualisation of
the structural features of the bacterial HslUV protein can be found at the PDB ‘Molecule of the Month’.
The structure of HslUV has provided insight into the structure of
proteasomes in general, as well as their mode of action.
Next: Table of AAA ATPases
Previous: Protein
Degradation in the Proteasome
