Importins
What InterPro Tells Us
Q14974 Human Importin, beta-1 subunit
InterPro Domain Architecture
![]()
InterPro Entry |
Signatures |
Graphical Match |
Method Name |
|
IPR000225 |
PF00514 |
|
Arm |
|
IPR000357 |
PF02985 |
|
HEAT |
|
IPR000357 |
PS50077 |
|
HEAT_REPEAT |
|
IPR001494 |
PF03810 |
|
IBN_N |
|
IPR001494 |
PS50166 |
|
IMPORTIN_B_NT |
|
IPR011989 |
G3DSA:1.25.10.10 |
|
ARM-like |
|
Structural Features |
|
|
|
|
1qgr |
1qgrA |
|
|
|
1.25.10.10.12 |
1qgrA0 |
|
|
|
a.118.1.1 |
d1ibrb_ |
|
|
From the graphical match in the table above, you can see that the signatures are grouped into four InterPro entries for human importin beta-1. These entries identify the domain architecture and sequence features of this protein.
DOMAIN Entries
Ø IPR001494: N-terminal domain of Importin, represented by two signatures: PF03810 (PFAM) and PS50166 (PROSITE).
Ø IPR011989: Armadillo-like helical domain, represented by one signature: G3DSA:1.25.10.10 (Gene3D).
REPEAT Entries
Ø IPR000225: Armadillo repeat, represented by three signatures: PF00514 (PFAM), PS50176 (PROSITE), and SM00185 (SMART) – although only one of these signatures (PF00514) hits this protein.
Ø IPR000357: HEAT repeat, represented by two signatures: PF02985 (PFAM) and PS50077 (PROSITE).
The importin beta-1 subunit consists of repetitive sequence elements. The armadillo repeat is a 40-amino acid repeated motif that can form an alpha-alpha-superhelical structure, and which was first identified in the Drosophila melanogaster segment polarity gene armadillo. Armadillo repeats function in a variety of processes, including intracellular signalling and cytoskeletal regulation, and occur in a wide variety of proteins. The HEAT repeat is related to the armadillo repeat, and consists of a 37-47-amino acid motif that forms an armadillo-like alpha-alpha-superhelical structure. HEAT repeats are found in a variety of cytoplasmic proteins, especially those involved in intracellular transport. The repeat entries IPR000225 and IPR000357 help to identify some of the several armadillo-like repeat motifs in this protein.
Entry IPR011989 classifies the importin protein as consisting of one armadillo-like superhelical domain. The IPR001494 entry identifies the N-terminal domain as having a separate, defined sequence motif.
Structural features
The remaining three entries in
the able above give information on the known and predicted structure of this
protein. These entries present known
structural data from the structural database PDB (green stripe) and the
structural classification databases CATH (pink stripe) and SCOP (black stripe)
(the names such as 1qgrA0 are derived from the PDB entry upon which
they are based, here PDB entry 1qgr, chain A).
The graphical match for the PDB entry 1qgr displays the full length of the original PDB entry, here
covering the entire protein. Both CATH
and SCOP classify this protein in the same way, namely as one armadillo-like
structural domain. The reason the SCOP
entry is shorter, is because the shorter PDB entry 1ibr is displayed for SCOP.
What the Structure Tells Us
Structures
associated with the human importin beta-1 subunit can be viewed using
AstexViewer®, which is linked from the Match Table via the logo ![]() on the InterPro page (please note, there is
no link directly from this page to the AstexViewer®, therefore you need to go
to the
on the InterPro page (please note, there is
no link directly from this page to the AstexViewer®, therefore you need to go
to the ![]() link on the InterPro page for Q14974). The AstexViewer® displays the PDB structure with the CATH and
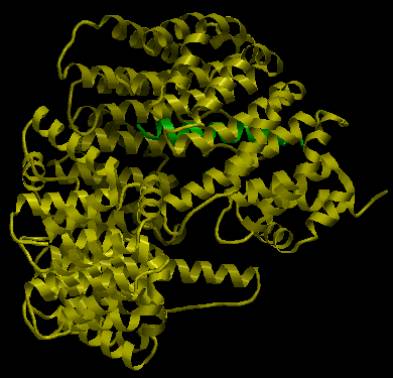
SCOP domains highlighted in yellow. In the
picture below, the CATH domain representing the alpha-alpha-superhelical
structure of the protein is shown.
link on the InterPro page for Q14974). The AstexViewer® displays the PDB structure with the CATH and
SCOP domains highlighted in yellow. In the
picture below, the CATH domain representing the alpha-alpha-superhelical
structure of the protein is shown.
|
|
|
AstexView of human importin beta-1 subunit: the alpha-alpha-superhelical topology forms one structural domain. |
There are structures available for
several importins from different organisms in the Protein Data Bank (PDB). A detailed description and visualisation of
the structural features of importins can be found at the PDB ‘Molecule of the Month’.
The crystallographic structures of various importins have provided insight
into the transport of molecules through the nuclear membrane.
Next: Table of Importin Proteins
Previous: Importing
Molecules into the Nucleus