Dionysian Mysteries The Aldehyde Dehydrogenase (aldh) Family
focusDo you have friends that cannot handle alcoholic drinks? Just half a pint of beer or a few sips of wine, and their faces turn red, possibly with some hangover symptoms, such as headaches and nausea? You may envy their cheap night out, but wonder why these people cannot tolerate alcohol as you do. The phenomenon is called ‘alcohol flush reaction’, also known as ‘Asian flush syndrome’, due to its association with the Asian population. It is a condition caused by the accumulation of acetaldehyde, a metabolic byproduct of the catabolic metabolism of alcohol.

Normally, during the alcohol metabolic process, ethanol is converted to acetaldehyde by an alcohol dehydrogenase enzyme, called ADH1B, and then broken down to acetic acid by an aldehyde dehydrogenase enzyme (ALDH).
In humans, there are nineteen identified ALDH genes (ALDH1-19). Most Europeans have normal copy of the ALDH2 gene, whilst approximately 30-50% of East Asians carry an allele (ALDH2*2) that results in the synthesis of a less efficient enzyme [1].
ALDH2 forms homotetramers. Each subunit in the tetramer consists of three domains - the catalytic domain, the coenzyme-binding domain and the oligomerisation domain. The low activity of ALDH2*2 is the result of a substitution of lysine for glutamate at position 487 (Glu487) of the 500-amino-acid mature enzyme [2]. The Glu487 links the coenzyme-binding site to the active site, which creates a stable structural scaffold contributing to catalysis (Figure 1). In the ALDH2*2 apoenzyme, the presence of a lysine at residue 487 disturbs the hydrogen bonds and causes disruptions of the αG helix structure [3]. This reduces affinity for the coenzyme and lowers the rate of the metabolic process [3].
As a result of this mutation, acetaldehyde accumulates whenever alcohol is consumed. Unfortunately, acetaldehyde is a DNA damaging agent that can cause cancer [4], and a higher risk of ALDH2-deficient drinkers developing esophageal cancer has been shown by several studies [4,5,6]. A knock out mouse model also links ethanol consumption with higher risk of acetaldehyde toxicity in ALDH2 deficient individuals [7]. But whilst the outlook seems to be dim and gloomy for the ALDH2*2 drinkers, on the bright side, they are less likely to suffer alcohol addiction problems [8]. In fact, there is a drug called disulfiram that causes symptoms similar to Asian flush syndrome that is used to treat alcoholism.
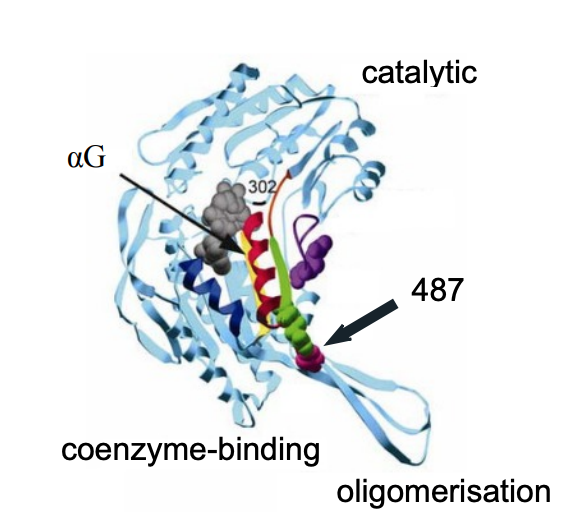 Figure 1. The protein structure of a single subunit of ALDH2.
Residue 487 is indicated in violet. ALDH2*2 αG helix is shown in red, wild type I shown in blue. Picture modified from Larson et al. 2005 [3].
Figure 1. The protein structure of a single subunit of ALDH2.
Residue 487 is indicated in violet. ALDH2*2 αG helix is shown in red, wild type I shown in blue. Picture modified from Larson et al. 2005 [3].
Interestingly, some ALDH2*2 individuals have less intense flushing symptoms. This is because they also have a less active form of ADH1B (ADH1B*1/*1). This prevents a steep rise in acetaldehyde after drinking. However, some studies have shown that these individuals may have higher risk of both alcoholism and cancer (Figure 2) [8, 9].
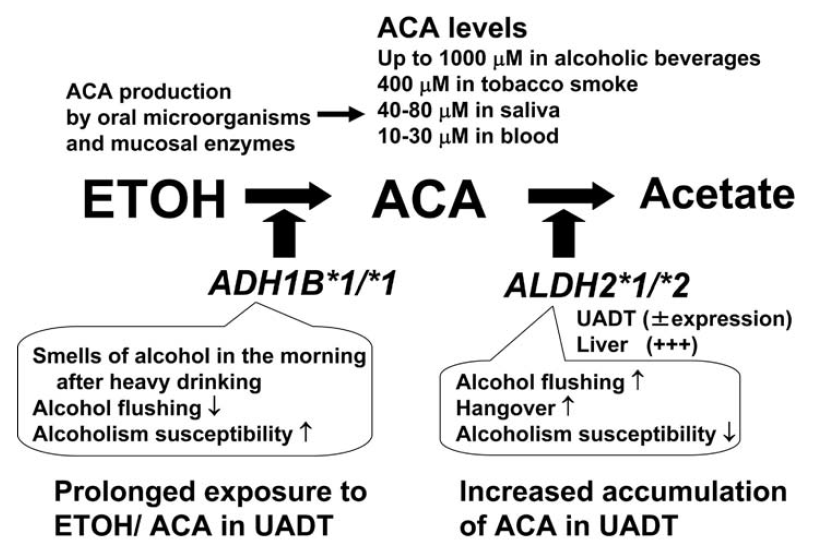 Figure 2. Ethanol metabolic process.
Figure 2. Ethanol metabolic process.
It is intriguing that a single mutation in the ALDH2 gene could cause alcohol-related health problems. From an evolutionary point of view, aldehyde dehydrogenases are utilised by different species to detoxify harmful chemical intermediates, and hence play an important role in cell survival. They catalyse the conversion of a wide variety of aldehyde substrates to their respective carboxylic acids, using coenzyme NAD or NADP. The aldehyde dehydrogenase family members contain two conserved sites: a cysteine active site and a glutamic acid active site (Figure 4). These two sites are represented by the InterPro entries IPR016160 and IPR029510, and are conserved across species, from archaea and bacteria to eukaryotes.
ALDH in different species
In contrast to humans, budding yeast have only five ALDHs. They are the key enzymes of the pyruvate dehydrogenase (PDH) bypass, which generates additional acetyl-CoA [10]. In the wine producing process, the acetate produced by the PDH bypass accumulates during the alcoholic fermentation of sugars [11]. The level of acetate has important effects on wine quality - most unspoiled wines have a level of 0.2 to 0.8 g of acetate per litre [12].
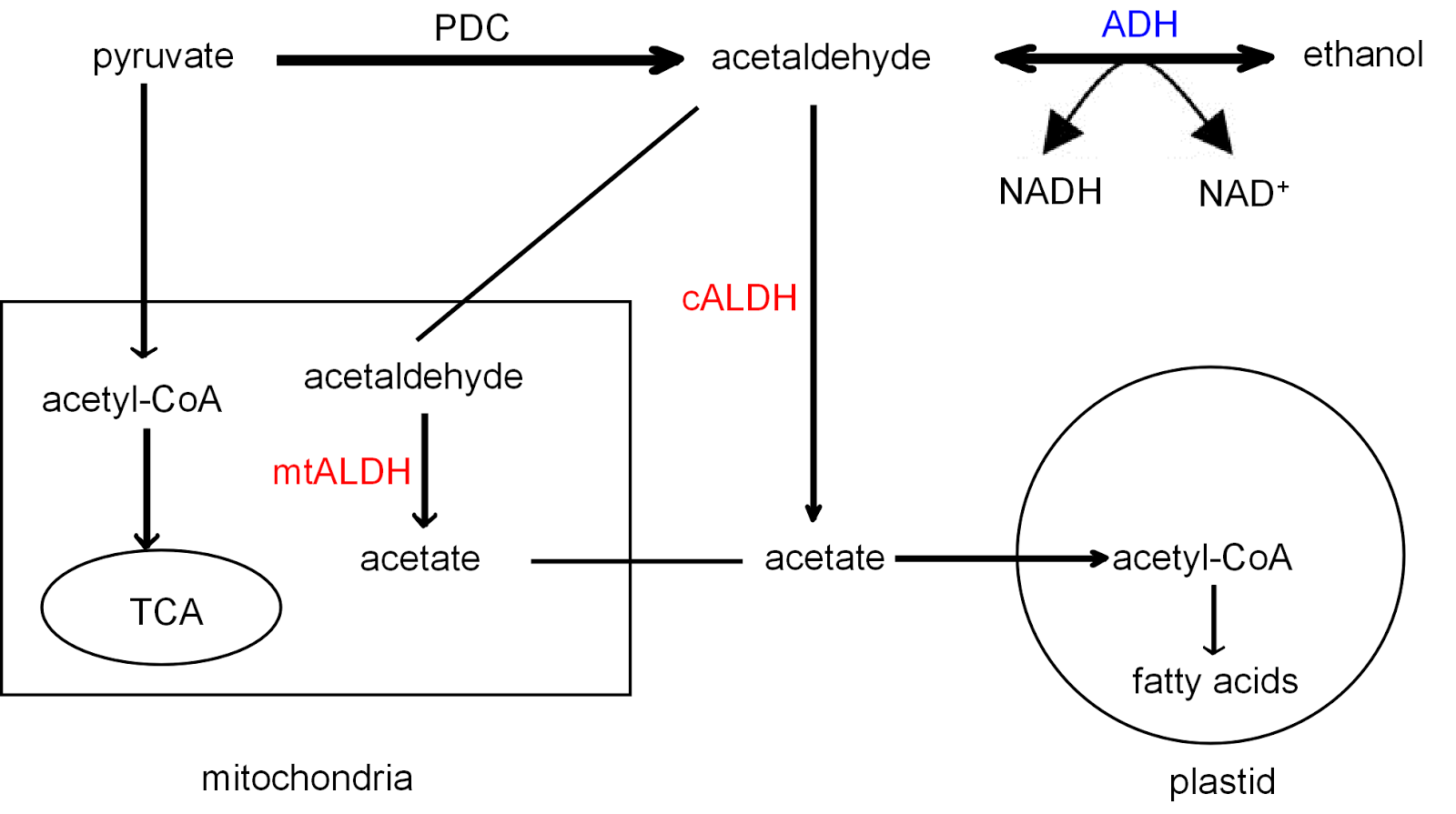 Figure 3. Key enzymes of the PDH bypass pathway. PDH, pyruvate
dehydrogenase; PDC, pyruvate decarboxylase; ADH, alcohol dehydrogenase; mtALDH, mitochondria ; cALDH, cytoplasmic ALDH. Modified from Wei
et al. 2009. [16]
Figure 3. Key enzymes of the PDH bypass pathway. PDH, pyruvate
dehydrogenase; PDC, pyruvate decarboxylase; ADH, alcohol dehydrogenase; mtALDH, mitochondria ; cALDH, cytoplasmic ALDH. Modified from Wei
et al. 2009. [16]
Plants also have multiple ALDHs, and 14 have been identified in Arabodopsis [13]. They play an important role in the adaptation of plants to various stresses, such as drought, salinity and extreme temperatures [14]. They may also be involved in different transduction pathways [15].
An unsolved mystery
We may not yet understand the reason why the ALDH2 deficiency is widespread in Asian populations. However, research can help us understand more about the relationship between ALDH2, cancers and alcoholism, as well potentially uncovering the safe number of alcohol units that ALDH2*2 individuals can consume.
So before you encourage your friends to have another glass of wine or a pint of beer, you may need to check if they have the Asian flush symptoms, or even review their ALDH2 phenotype!
References
-
Helminen A, Väkeväinen S, Salaspuro M. ALDH2 genotype has no effect on salivary acetaldehyde without the presence of ethanol in the systemic circulation. PLoS One. 8(9):e74418. 2013. [PMID: 24058561]
-
Larson HN, Zhou J, Chen Z, Stamler JS, Weiner H, Hurley TD. Structural and functional consequences of coenzyme binding to the inactive asian variant of mitochondrial aldehyde dehydrogenase: roles of residues 475 and 487. 282(17):12940-50. J Biol Chem. 2007. [PMID:17327228]
-
Larson HN, Weiner H, Hurley TD. Disruption of the coenzyme binding site and dimer interface revealed in the crystal structure of mitochondrial aldehyde dehydrogenase “Asian” variant. J Biol Chem. 280(34):30550-6. 2005. [PMID: 15983043]
-
Lewis SJ, Smith GD. Alcohol, ALDH2, and esophageal cancer: a meta-analysis which illustrates the potentials and limitations of a Mendelian randomization approach. Cancer Epidemiol Biomarkers Prev. 14(8):1967-71. 2005. [PMID: 16103445]
-
Yokoyama A, Omori T, Yokoyama T. Alcohol and aldehyde dehydrogenase polymorphisms and a new strategy for prevention and screening for cancer in the upper aerodigestive tract in East Asians. Keio J Med. 59(4):115-30. 2010. [PMID: 21187698]
-
Seitz HK, Meier P. The role of acetaldehyde in upper digestive tract cancer in alcoholics. Transl Res. 149(6):293-7. 2007. [PMID:17543846]
-
Isse T, Oyama T, Matsuno K, Ogawa M, Narai-Suzuki R, Yamaguchi T, Murakami T, Kinaga T, Uchiyama I, Kawamoto T. Paired acute inhalation test reveals that acetaldehyde toxicity is higher in aldehyde dehydrogenase 2 knockout mice than in wild-type mice. J Toxicol Sci. 30(4):329-37. 2005. [PMID: 16404141]
-
Yokoyama A, Omori T, Yokoyama T. Alcohol and aldehyde dehydrogenase polymorphisms and a new strategy for prevention and screening for cancer in the upper aerodigestive tract in East Asians. Keio J Med. 59(4):115-30. 2010. [PMID: 21187698]
-
Lee CH, Lee JM, Wu DC, Goan YG, Chou SH, Wu IC, Kao EL, Chan TF, Huang MC, Chen PS, Lee CY, Huang CT, Huang HL, Hu CY, Hung YH, Wu MT. Carcinogenetic impact of DH1B and ALDH2 genes on squamous cell carcinoma risk of the esophagus with regard to the consumption of alcohol, tobacco and betel quid. Int J Cancer. 122(6):1347-56. 2008. [PMID:18033686]
-
Boubekeur S, Camougrand N, Bunoust O, Rigoulet M, Guérin B. Participation of acetaldehyde dehydrogenases in ethanol and pyruvate metabolism of the yeast Saccharomyces cerevisiae. Eur J Biochem. 268(19):5057-65. 2001. [PMID: 11589696]
-
Saint-Prix F, Bönquist L, Dequin S. Functional analysis of the ALD gene family of Saccharomyces cerevisiae during anaerobic growth on glucose: the NADP+-dependent Ald6p and Ald5p isoforms play a major role in acetate formation. Microbiology. 150(Pt 7):2209-20. 2004. [PMID: 15256563]
-
Remize F, Roustan JL, Sablayrolles JM, Barre P, Dequin S. Glycerol overproduction by engineered saccharomyces cerevisiae wine yeast strains leads to substantial changes in Byproduct formation and to a stimulation of fermentation rate in stationary phase. Appl Environ Microbiol. 65(1):143-9. 1999. [PMID: 9872772]
-
Kirch HH, Schlingensiepen S, Kotchoni S, Sunkar R, Bartels D. Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant Mol Biol. 57(3):315-32. 2005. [PMID: 15830124]
-
Zhang Y, Mao L, Wang H, Brocker C, Yin X, Vasiliou V, Fei Z, Wang X. Genome-wide identification and analysis of grape aldehyde dehydrogenase (ALDH) gene superfamily. PLoS One. 7(2):e32153. 2012. [PMID: 22355416]
-
Kirch HH, Bartels D, Wei Y, Schnable PS, Wood AJ. The ALDH gene superfamily of Arabidopsis. Trends Plant Sci. 9(8):371-7. 2004. [PMID: 15358267]
-
Wei Y, Lin M, Oliver DJ, Schnable PS. The roles of aldehyde dehydrogenases (ALDHs) in the PDH bypass of Arabidopsis. BMC Biochem. 10: 7. 2009. [PMID: 19320993]
