Homologous superfamily: a new InterPro entry type
interproInterPro entries are comprised of one or more signatures from member databases, thereby providing a unified view of protein families. Each InterPro entry is labelled with a “type” reflecting what the constituent signatures represent: families, domains, repeats or sites. As part of release 65.0, InterPro has added a new entry type, homologous superfamily, to complement the existing set of types. A homologous superfamily is a group of proteins that share a common evolutionary origin, indicated by their structural similarities.
Why create a new entry type?
We have introduced the homologous superfamily type to accommodate signatures from CATH-Gene3D and SUPERFAMILY. Both of these databases use a slightly different methodology to InterPro’s other member databases, in that they often utilise a collection of underlying profile hidden Markov models (HMMs) to represent diverse structural families, rather than one single model. Integrating these multi-profile-HMM signatures into InterPro entries alongside our other member database signatures has not always been straightforward, as they tend to match wider sets of proteins. As a result, we have not been able to include as much information from these databases as we would like. Creating the new entry type better encapsulates what CATH-Gene3D and SUPERFAMILY are trying to describe and allows us to better represent their data in InterPro.
The last CATH-Gene3D update within InterPro highlighted this aforementioned discordance between signature generation. In this particular release of CATH-Gene3D, many of their overarching signatures had been substantially improved with both new and updated profile HMMs, to capture more diverse members of the families. While this increase in sequence coverage is superb, InterPro entries containing CATH-Gene3D signatures with other member databases (such as Pfam) no longer represented the same protein family, and so had to be separated.
How do we represent the new entry type?
Homologous superfamily entries are exclusively composed of CATH-Gene3D and/or SUPERFAMILY methods. They are not arranged in hierarchies, and exist as separate entities. A typical homologous superfamily entry is shown in Figure 1.
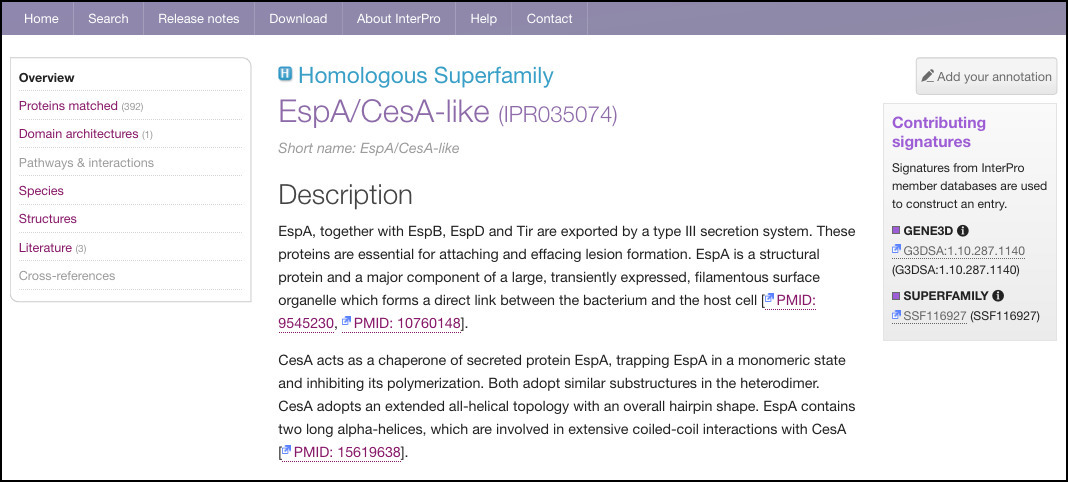 Figure 1. InterPro entry page for homologous superfamily EspA/CesA-like.
Figure 1. InterPro entry page for homologous superfamily EspA/CesA-like.
Within the protein overview page ‘Homologous superfamilies’ matches are shown on a separate track, in the same way as ‘Domain and repeats’ (see Figure 2). This view shows the relationships between the different types in InterPro.
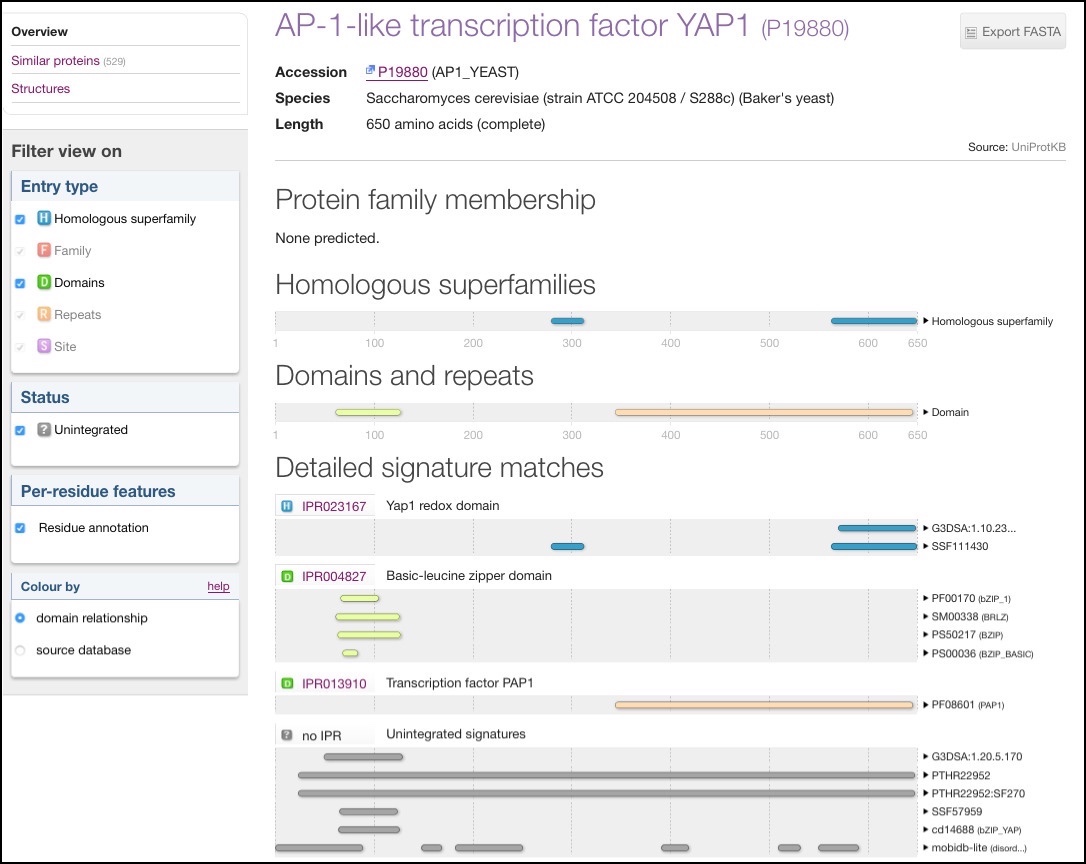 Figure 2. InterPro protein overview page for AP-1-like transcription factor YAP1 (P19880) showing homologous superfamily match.
Figure 2. InterPro protein overview page for AP-1-like transcription factor YAP1 (P19880) showing homologous superfamily match.
What does this change mean in terms of sequence analysis?
In the short term, this change will have little effect on sequence analysis using InterPro. All member database matches will be calculated as before, but existing CATH-Gene3D and SUPERFAMILY methods have been removed from existing InterPro entries and reintegrated into new entries corresponding to the homologous superfamily type. Over time, we anticipate that the change will allow us to integrate many more entries from these databases, increasing the sequence and amino acid residue coverage provided by InterPro, and allowing us to better represent structural-based signatures. We will also develop automated approaches for linking between homologous superfamilies and other entry types (such as families or domains) to allow the connection of structural data to other entries within InterPro that may lack a sequence representative with a known protein structure.
