Introducing the new interpro website
interpro
We released a new version of the InterPro website in September 2019. This release was a complete ground-up redesign of the website involving changes to both the underying data and the web interface. Given the scale of changes we made, perhaps it’s unsurprising that we encountered some hiccups. We’ll talk about those later in this post.
Goals
Add commonly requested features
We collated user-requests (from helpdesk emails), and internal team suggestions into a list of the most commonly requested website features.
Provide member database entry pages
One of our key goals was to improve access to InterPro member database information within the new website. For example, not all signatures from member databases have been integrated into InterPro entries yet. We wanted to improve the exposure of member database entries, regardless of intergration in InterPro, in the new site.
Ensure codebase is flexible
The technologies and code used to build the previous version of the website were becoming increasingly outdated, limiting the addition of new features to the website. We wanted to ensure the future adaptability of the website.
Improve scalability of InterPro releases
InterPro is currently updated approximately every 8 weeks. The ever-increasing volumes of sequence data submitted to public resources meant that the various calculations required for the release process were beginning exceed the time available to perform them. We wanted to optimize the release process to permit futher future scalability.
New features
We’ll be going into more detail about the new website and set-up in future posts, but it’s worth highlighting some key additions which we hope improve the experience of using InterPro.
Addition of new browsable data types: taxonomy, proteome, and set (from Pfam and Panther member databases)
The addition of new datatypes can be seen throughout the site, but the browse tab is a good starting point to see how we have connected these datatypes to our InterPro data.

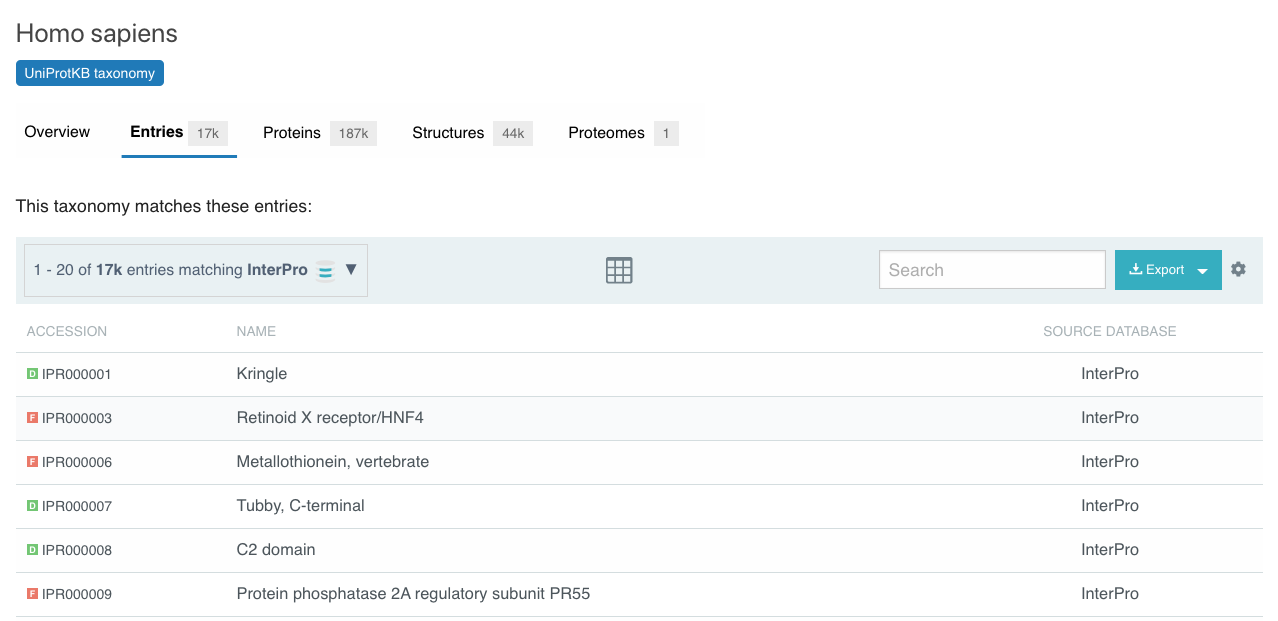
Ability to view/download entries matching a specific proteome or taxonomic node
The new taxonomy and proteome pages provide lists of all entries, proteins and structures related to that taxonomic node or proteome. This can be used to explore data at different levels of the taxonomic tree.
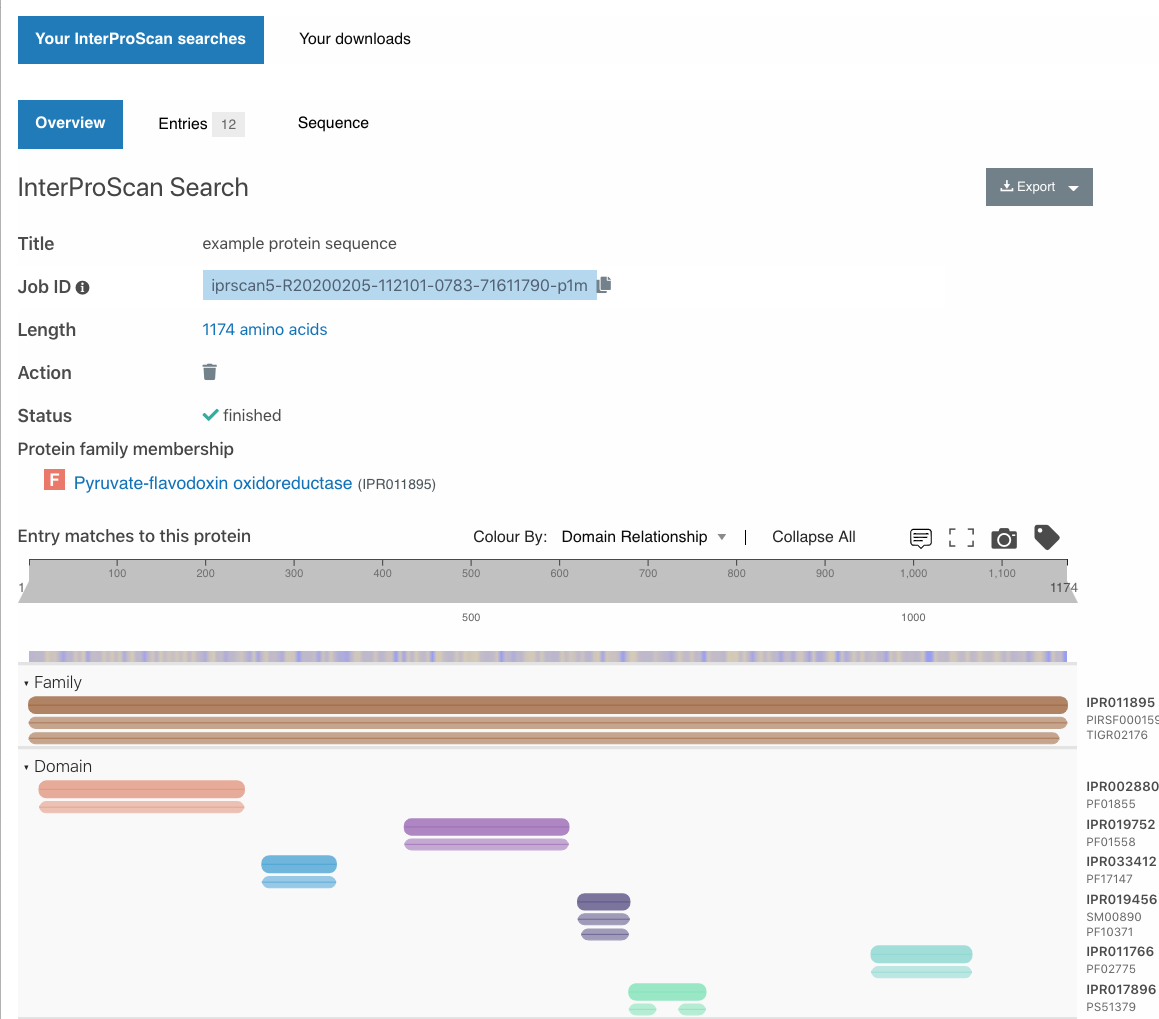
Ability to view InterProscan results within the website
As with the previous version of the InterPro website, InterProScan sequence searches can be submitted from the search page. It is now possible to view the results within the website. We have added the ability to view and share the results for as long as they are stored in the InterProScan service (currently results are stored on our servers for 1 week).
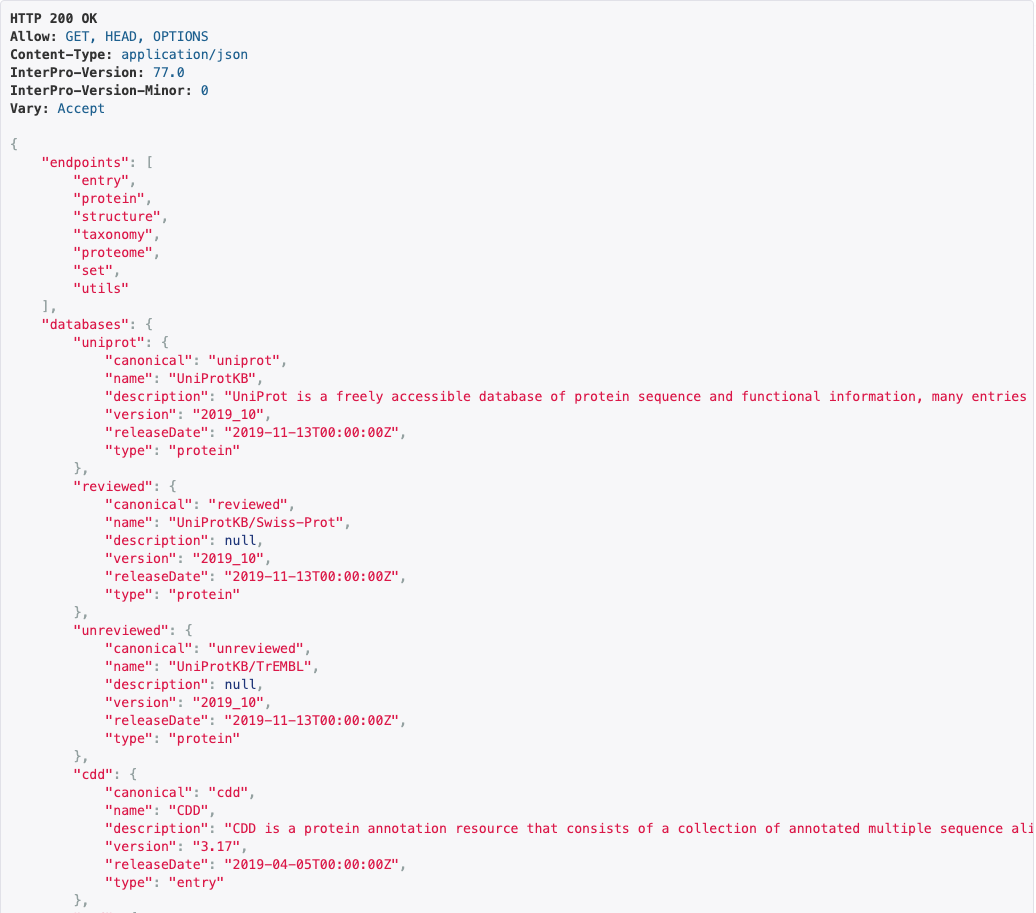
Introduction of a new Application Programming Interface (API) for downloading data via scripts
The new website actually makes use of the API to provide the content of the various webpages. The data can also be accessed directly without a browser. Documentation for the API can be found at our github repository
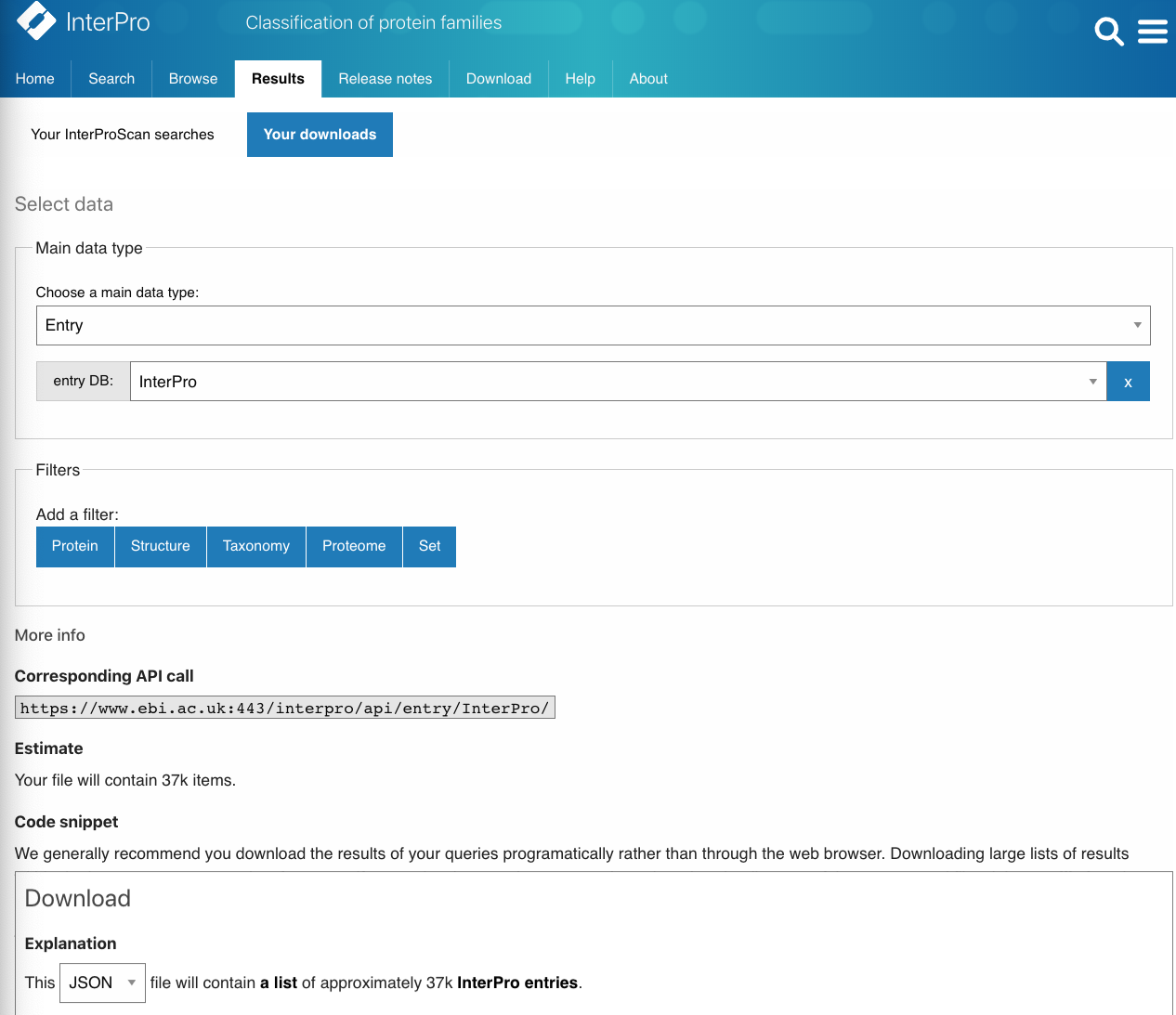
Automated script generator to help get started with programmatic access.
The automated script generator is a great starting point for using our API programmatically as it creates scripts in one of three languages, Python, Javascript or Perl. These scripts can be can be edited to run more complex queries.
InterPro member database entry pages searchable and viewable throughout the website
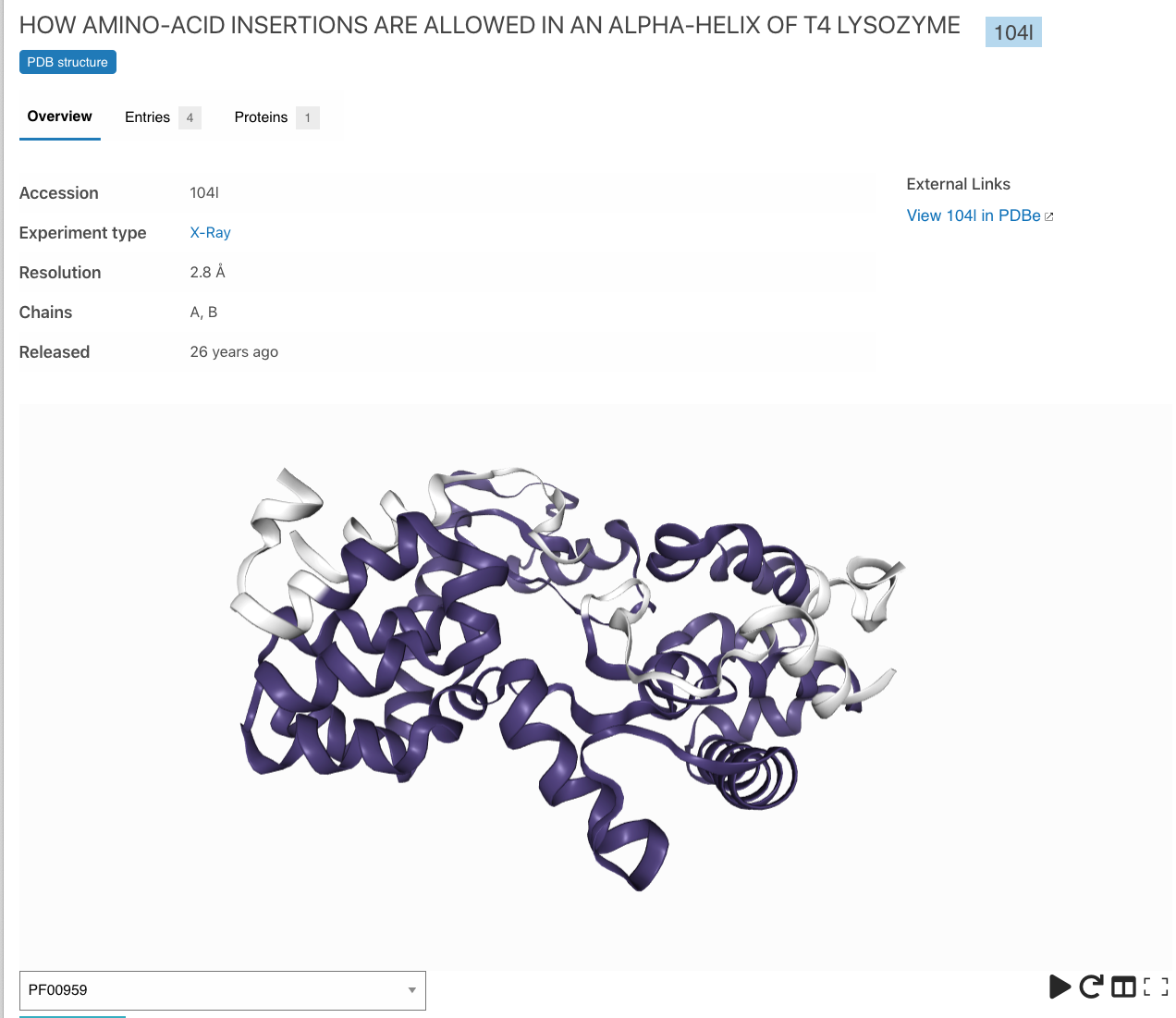
Interactive 3D representations of structures, highlighting the location of entry matches within the structure.
New Nightingale graphical view of protein shows the following information:
- Hydrophobicity plot of whole protein.
- Match convservation plot of Pfam matches to protein, showing the extent to which the protein matches the HMM model.
- Genome 3D predictions on protein (where available).
- Isoforms can be selected to view matches to particular splice variants.
Modern responsive design
Integration of modern web standards to provide improved user experience on a variety of devices, and increase the discoverability of our data through search engines e.g BioSchema.
Modular components
Some elements of the new website are available as custom elements, and can be downloaded and integrated into users own tools and websites.
Problems and roadmap
As we mentioned earlier, the launch of the new site has not been without issues. Many of these have been due to the processing load on the site, and would normally be resolved by the addition of more hardware capacity. Unfortunately the addition of more hardware has been delayed due to a pending relocation of the EBI data centre.
In the meantime we’ve been working to try to make the site more efficient by caching website queries for rapid response when the same query is made again. We have also been assessing queries that cause particularly heavy load on the servers, and pre-computing the data for those queries where possible.
Another major aspect of the work we’ve been focussing on has been server efficiency. We’ve been looking into optimising threading and connection management to get the best performance out of the hardware we have. We have also been investigating running parallel clusters of nodes to allow us to increase the number of requests we can handle simulataneously, although this work is still at an experimental stage. We have also moved our databases onto solid-state drives (SSD) to reduce general query times.
We will continue to monitor site performance and make improvements until we have the website working faster. As mentioned earlier, we should be able to add extra hardware to our website and API once the data centre relocation has been completed.
In addition to this we have been optimising the user experience side of things and responding to requests to enable users to get to the data that they want as efficiently as they can. With this in mind we have also been undertaking a small User Experience (UX) study to see where we should apply our efforts.
We’ll keep you updated with more information soon.
