New features in InterPro 82.0
interproInterPro 82.0 has been released with plenty of new InterPro entries, but also with many new features and improvements. This blog post describes the new key features you can expect to encounter while navigating the InterPro website.
Pagination of structure models in the structure section for proteins
Protein structure models from CATH and SCOP databases are displayed in the Structure subsection of protein pages in the protein viewer under the “Features” section. Until now, all the models were shown, making the display too busy for proteins with a lot of models (e.g. Q04049). We have changed the way this is handled by adding pagination for this section in the viewer, displaying 10 models at a time. The user can look at the following/previous models by clicking on the Previous or Next buttons respectively.
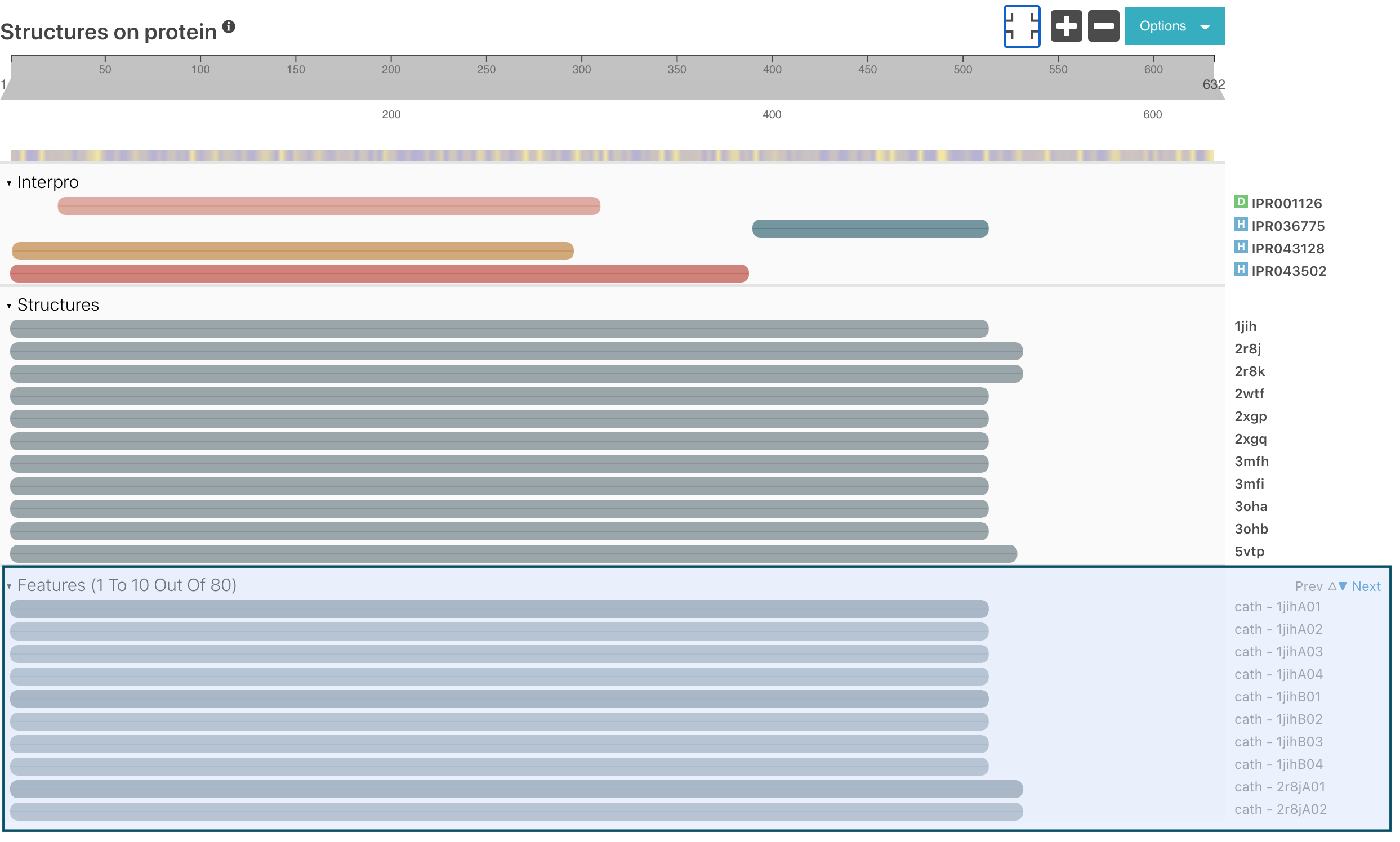 Figure 1. Features section with pagination for structural models matching Q04049.
Figure 1. Features section with pagination for structural models matching Q04049.
Entries can be tagged as Favourites
Users can now save their favourites entries and access them directly from the InterPro Home page. To add a favourite entry, the user needs to click on the Star symbol next to the InterPro entry name in the InterPro entry page. The star will then turn from grey to orange. The list of pinned entries is accessible from the Home page in the Favourite tab located next to the Latest entries tab in the data section.
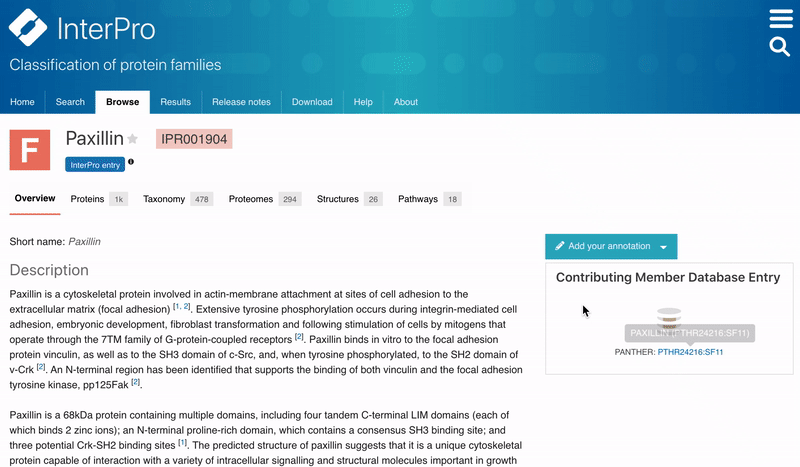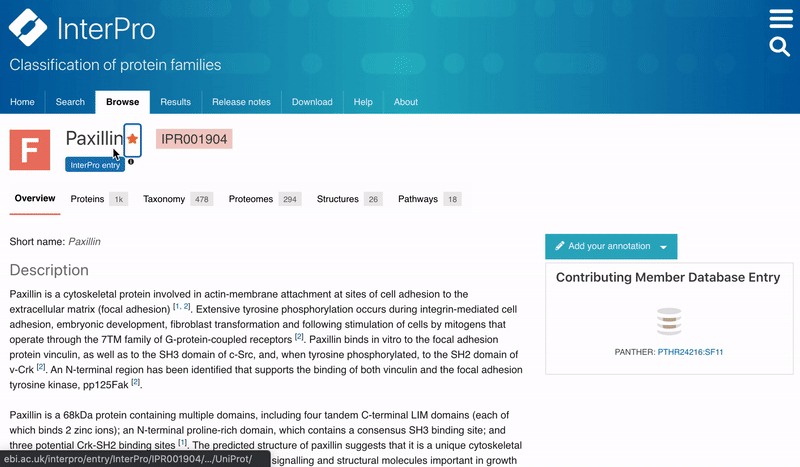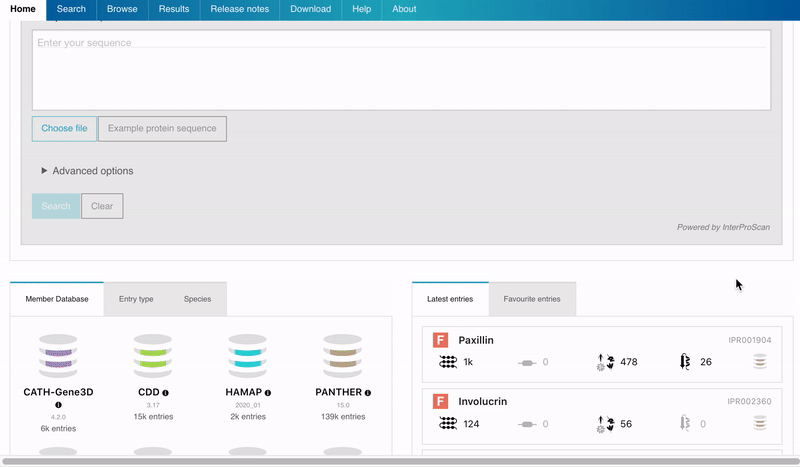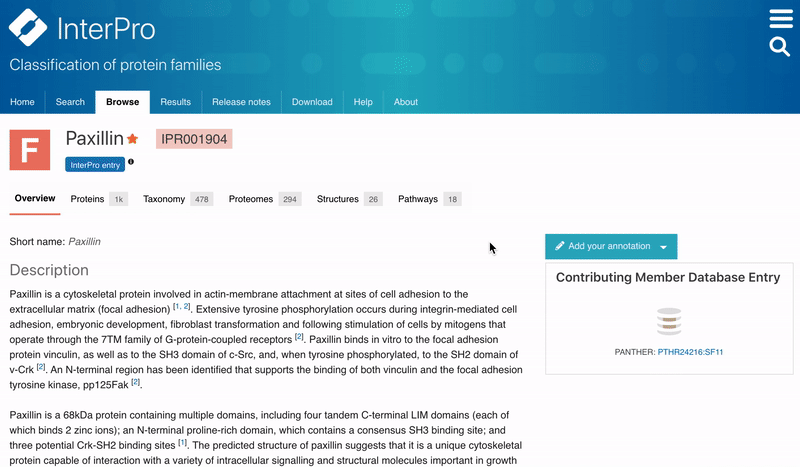 Figure 2. Adding a favourite entry and accessing the list of Favourites
Figure 2. Adding a favourite entry and accessing the list of Favourites
Pfam signature page updates
Two new tabs are available in the Pfam member database pages: Domain architectures and Curation.
Domain Architectures
The Domain Architectures subsection displays the different domains arrangements for all the proteins matching the Pfam signature being looked at.
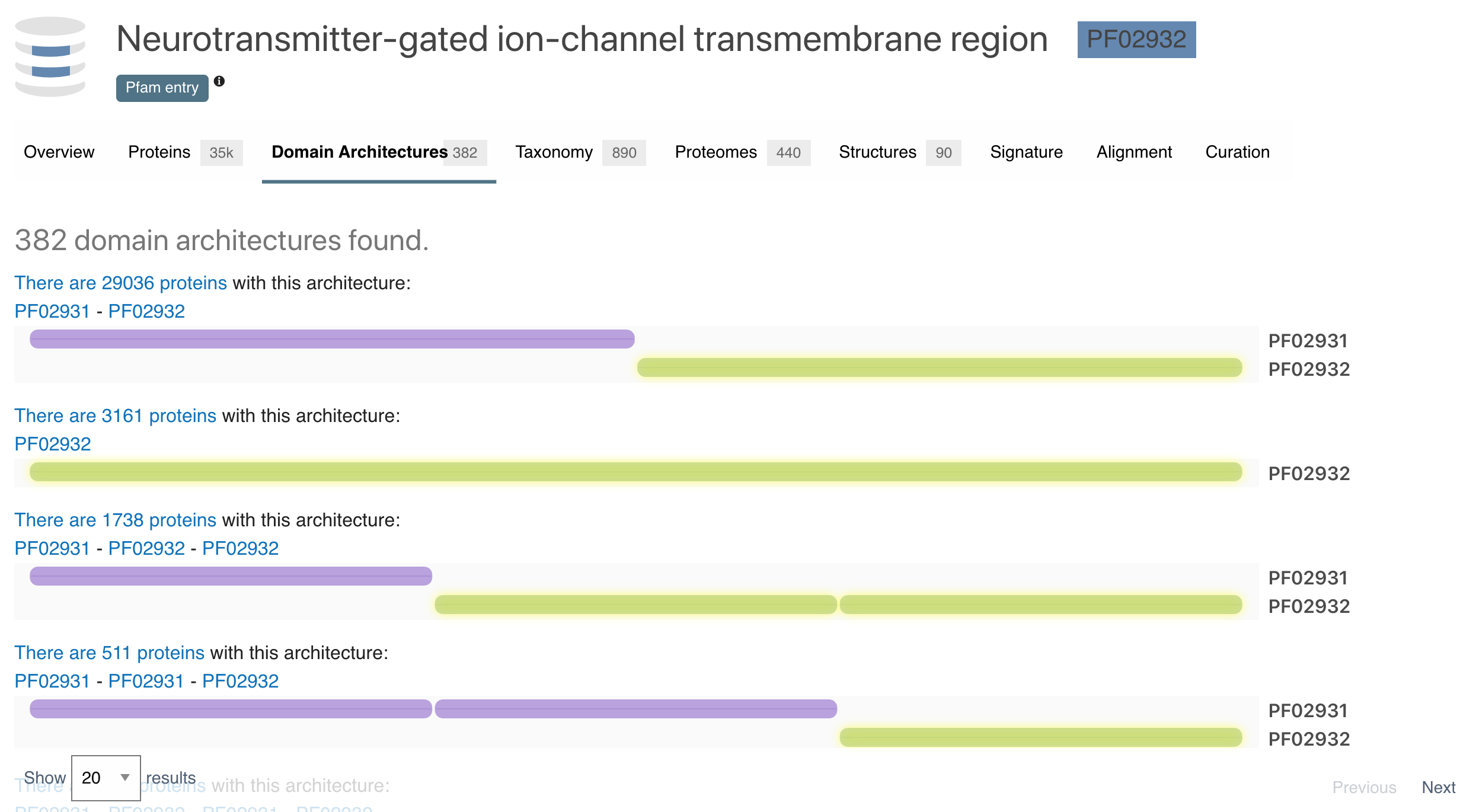 Figure 3. 4 out of 382 different domain architectures for the proteins matching Pfam signature PF02932.
Figure 3. 4 out of 382 different domain architectures for the proteins matching Pfam signature PF02932.
Curation
The Curation subsection of a Pfam signature displays information about the Pfam curators who annotated the signature, Sequence ontology term and HMM characteristics. Downloading the HMM file is also possible from this tab.
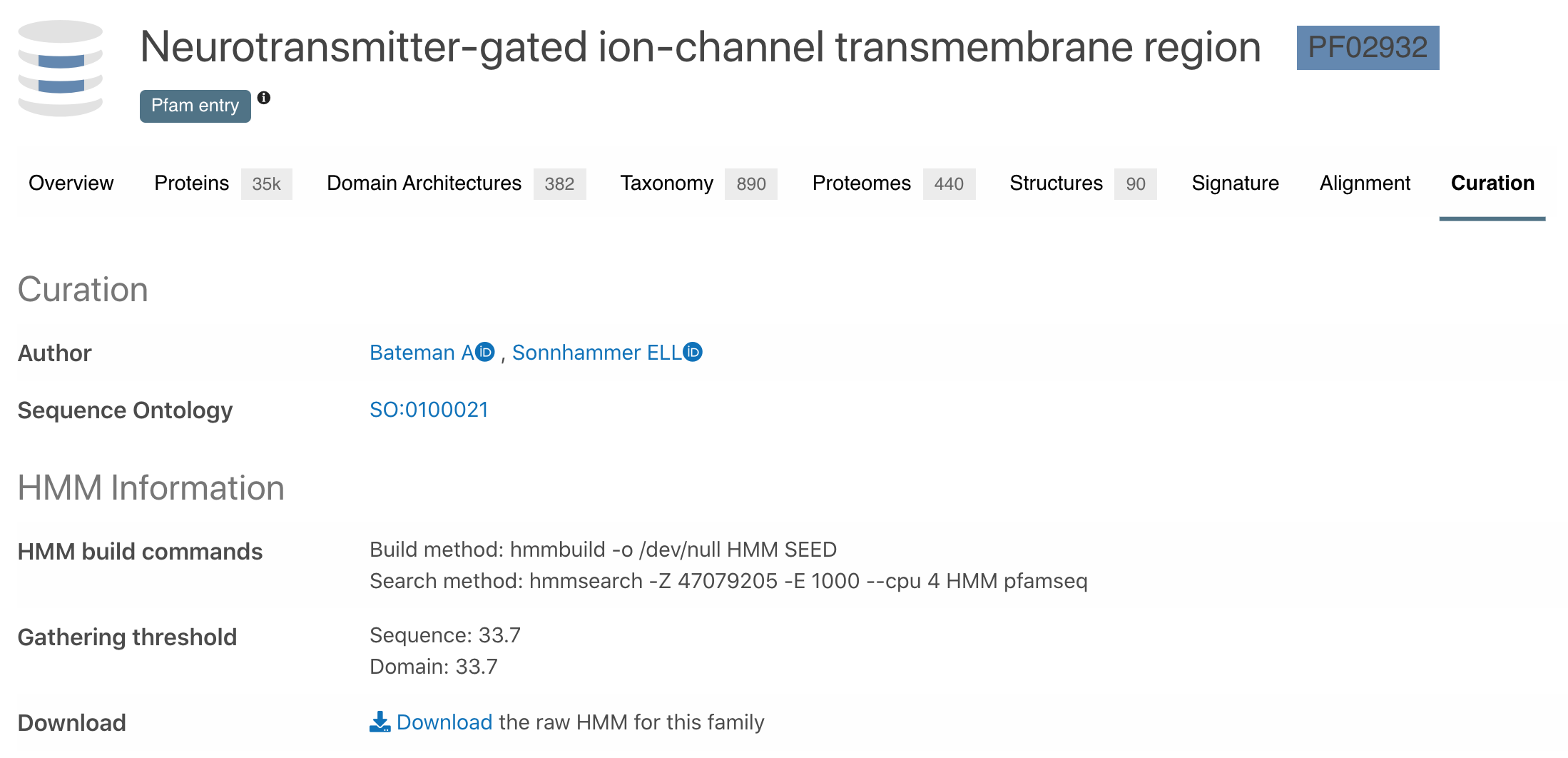 Figure 4. Curation subsection for the Pfam signature PF02932.
Figure 4. Curation subsection for the Pfam signature PF02932.
Wikipedia information
Wikipedia information has been added to the Pfam signature Overview section, when available, providing additional description concerning the signature. Because Wikipedia articles can be extremely variable in length we decided to only show the first paragraph of the Wikipedia page, clicking on the section title opens the full Wikipedia page.
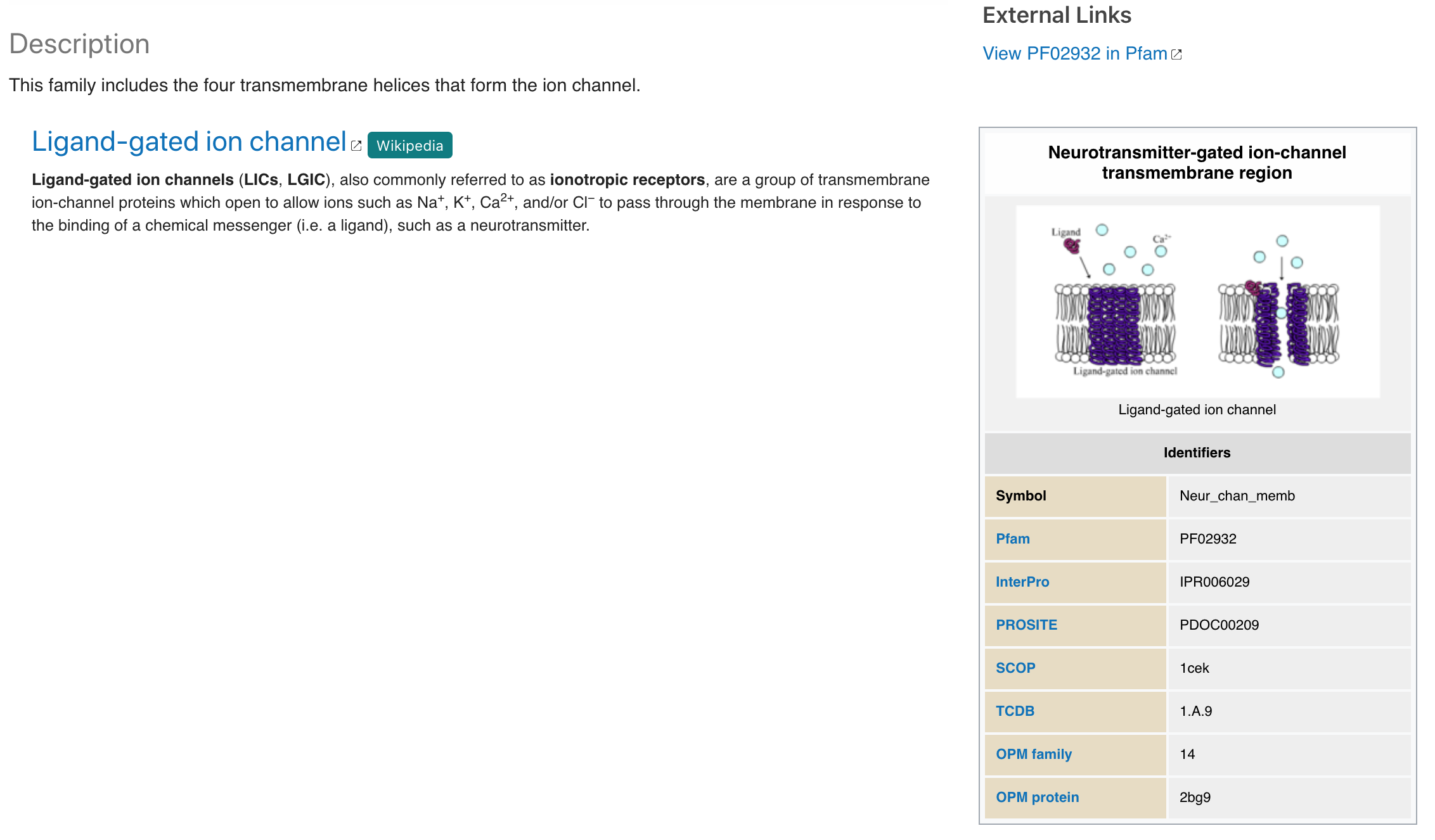 Figure 5. Wikipedia section for Pfam signature PF02932.
Figure 5. Wikipedia section for Pfam signature PF02932.
Literature references in Pfam sets
Literature references are now displayed in Pfam sets overview pages. This data gives more complete information to the description of the set.
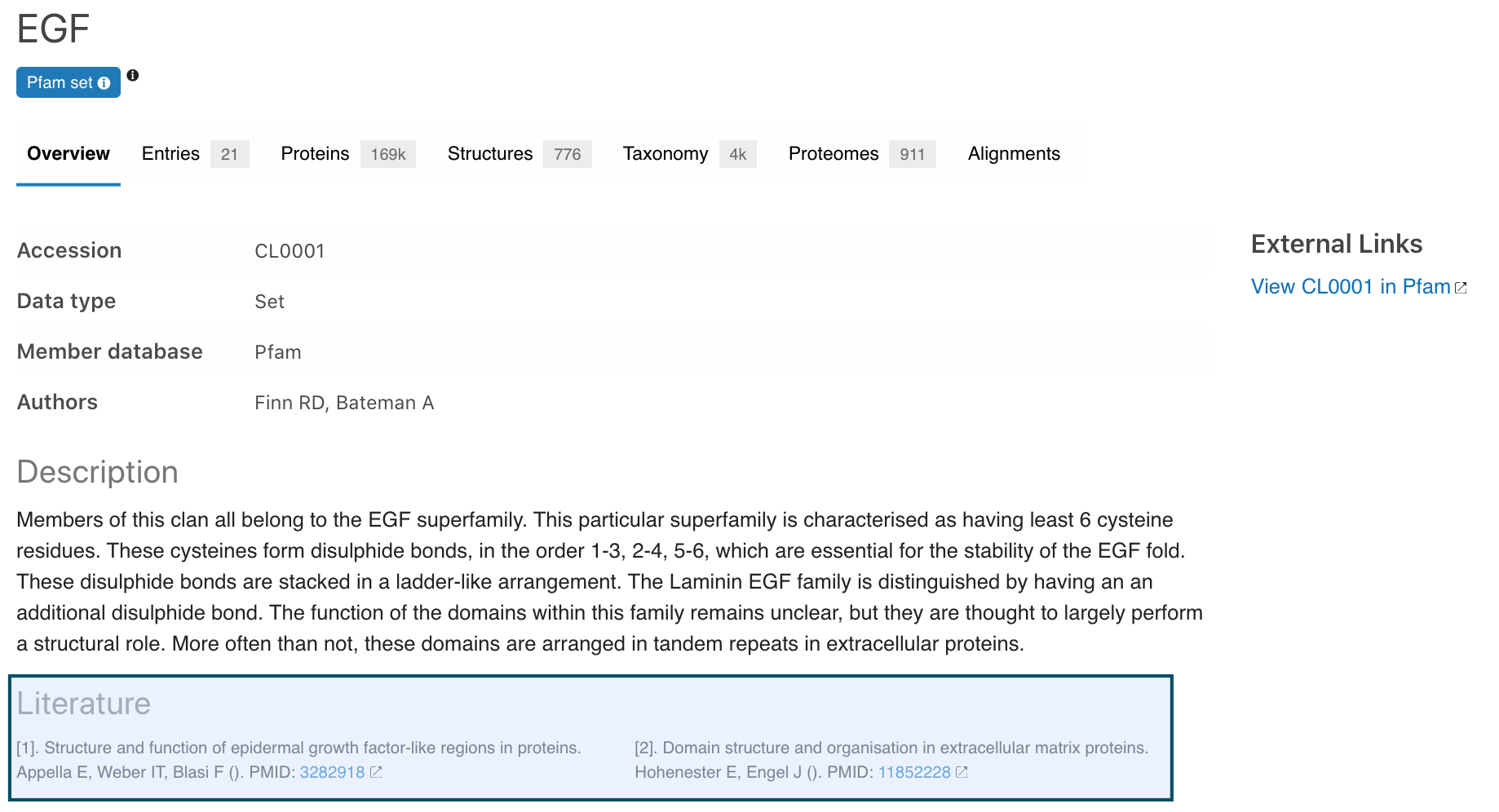 Figure 6. Literature information for Pfam set CL0001.
Figure 6. Literature information for Pfam set CL0001.
Enable/Disable searched term highlight
When performing a text search, the searched term is highlighted by default in the search results. A new button has been added to Enable/Disable the highlight, it is located next to the Export button (see figure below). The choice is saved in the session and reflected in other pages as well.
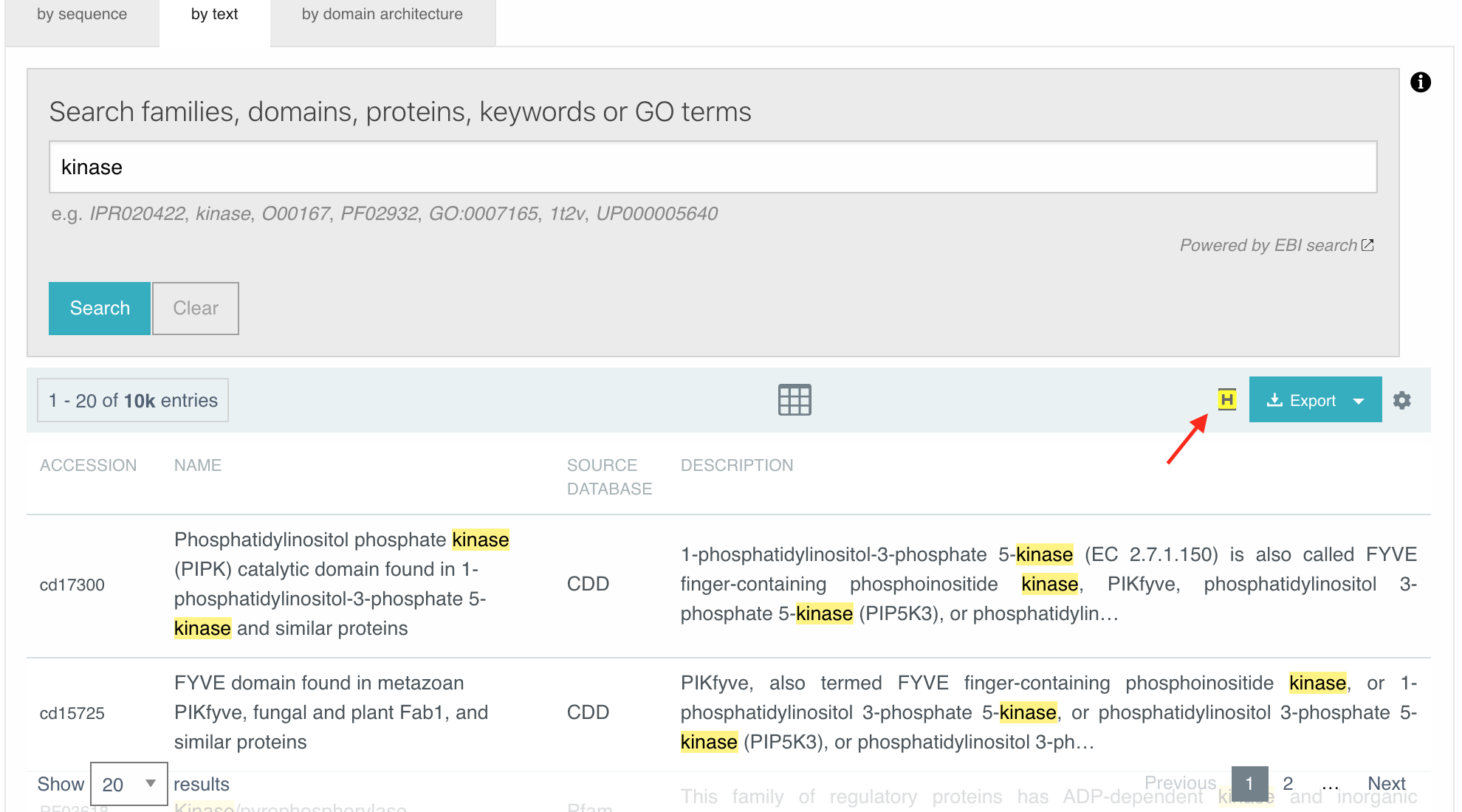 Figure 7. Highlight of the “kinase” text search term in the list of results. The highlight can be disabled by clicking on the H button (pointed by the red arrow).
Figure 7. Highlight of the “kinase” text search term in the list of results. The highlight can be disabled by clicking on the H button (pointed by the red arrow).
