Interpro 83.0 Key Features
interproInterPro 83.0 has been released with plenty of new InterPro entries, but also with many new features and improvements. This blog post describes the new key features you can expect to encounter while navigating the InterPro website.
Protein entry page
A protein entry page contains information on a specific protein provided by UniProt.
Isoform identifier in URL
The protein entry page offers the possibility to display the InterPro matches for different isoforms of a protein in the sequence viewer when they exist. The selected isoform is now exposed in the URL (e.g. https://www.ebi.ac.uk/interpro/protein/reviewed/O00167/?isoform=O00167-2). Split comparison mode for isoforms in full screen The protein isoform selected matches in the sequence viewer can be compared to the default protein matches in full screen mode. To do so click on the split icon located next to the isoform selection drop down menu once an isoform has been chosen, as shown in Figure 1.
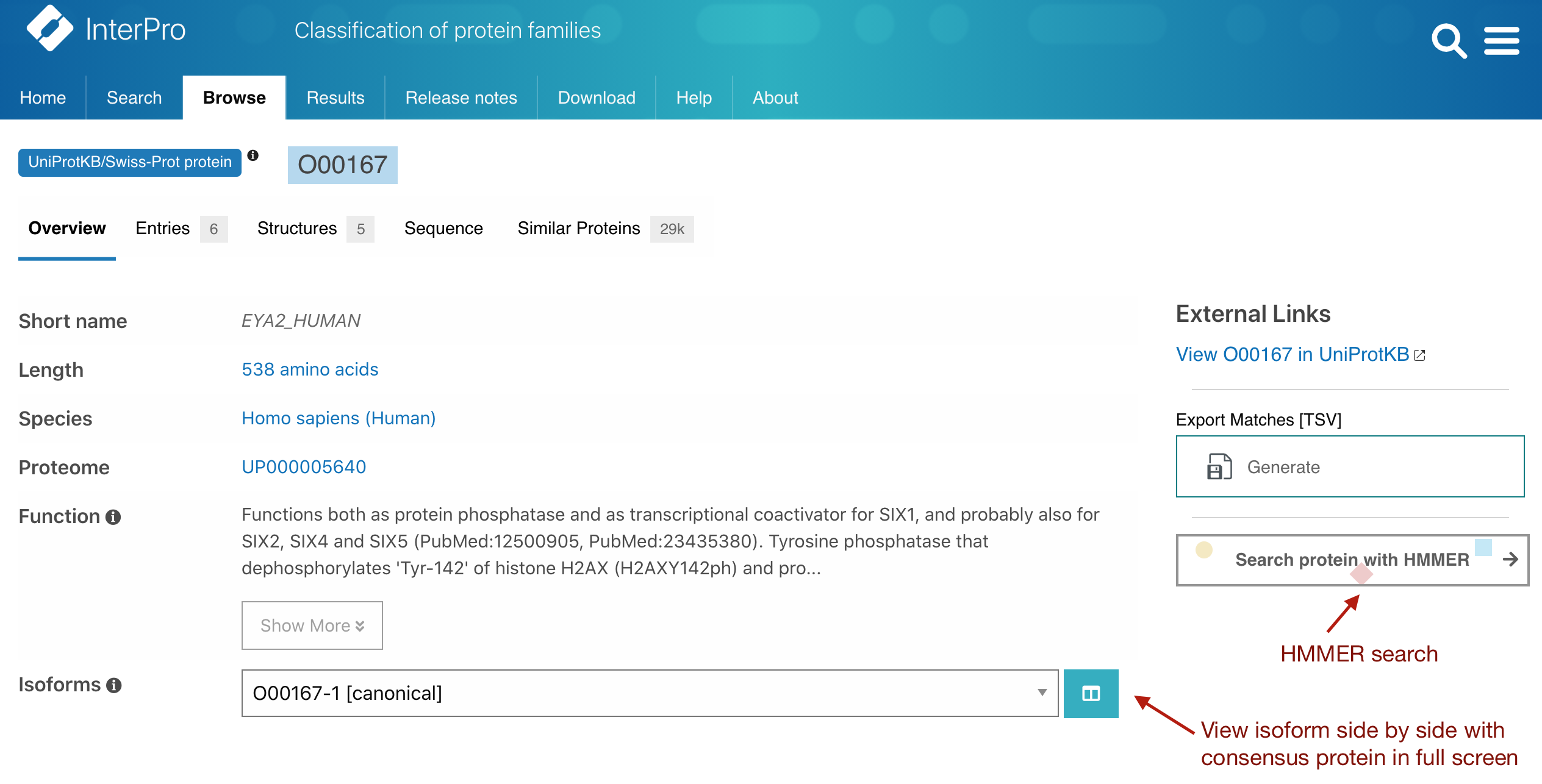 Figure 1. Split comparison view and HMMER search from the InterPro protein entry page for O00167.
Figure 1. Split comparison view and HMMER search from the InterPro protein entry page for O00167.
HMMER search feature
The protein entry page offers the possibility to perform a HMMER search with the selected protein. This can be done by clicking on the “Search protein with HMMER” button located on the right hand side of the page, as shown in Figure 1. Alternatively, you can go to the Sequence tab and select a section of the sequence to start your search.
Taxonomy subpage
The taxonomy subpage is available under the InterPro Entry, Member database and Set entry pages. It provides the list of species the entity is matching, based on data from UniProt taxonomy.
Customisable display
Previously, the taxonomy subpage was displaying key species matches and below it all the matching organisms. You can now choose to display the key species, the organisms table or both by selecting the corresponding option in the InterPro settings. This allows the display of unwanted data and improves the performances. Settings are accessible from the InterPro website header under the hamburger icon or when there is a wheel icon on a table header.
Accessing extra information
Links to the matching proteins, taxonomy information and the fasta file generating option have been grouped together in the key species and organisms tables in the ACTIONS column (see Figure 2), making the access to additional information clearer. Similar data access has also been added for Proteomes subpages.
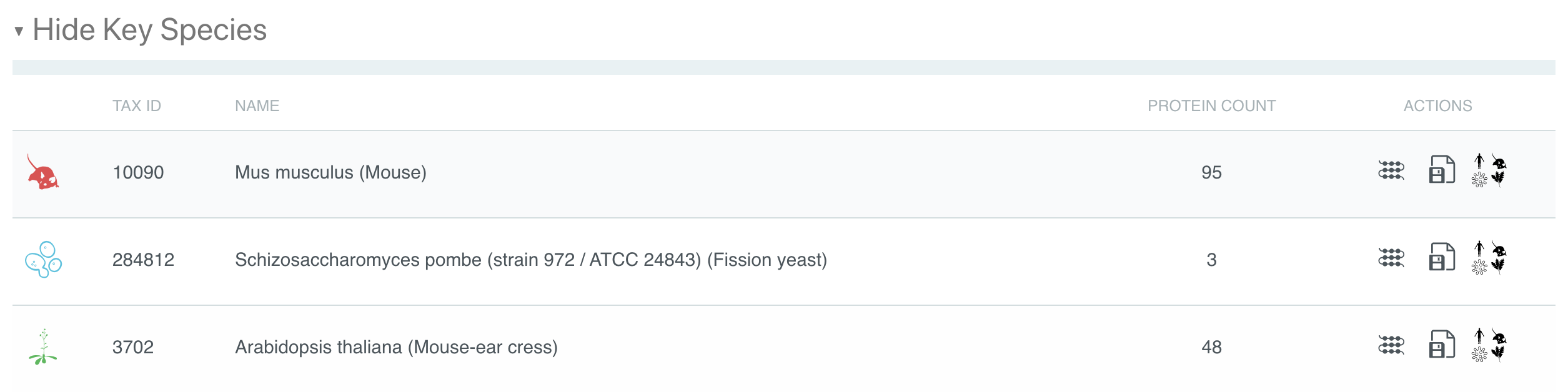 Figure 2. Taxonomy subpage key species table actions for IPR020422.
Figure 2. Taxonomy subpage key species table actions for IPR020422.
Visualising InterProScan output files
InterProScan, the underlying software behind the InterPro protein sequence search can be downloaded and ran using the command line. Input sequences, which can be nucleotide or protein sequences are submitted in FASTA format. Matches are then calculated against all of the requested member database’s signatures and the results are then output in a variety of formats (TSV, XML, JSON, GFF3, HTML, SVG). The InterProScan output files in JSON format can now be imported into the InterPro website to visualise the results, providing a graphical view of the matches of each protein sequence, similar to the results of a protein sequence search performed on the website. If InterProScan was run using nucleotide sequences, a job result is created for each Open Reading Frame (ORF) and ORFs from the same nucleotide sequence are grouped accordingly. This is available under the Results page Your InterProScan searches section by clicking on the import icon located on the right hand side of the page, above the jobs table, as shown in Figure 3 below.
This import feature can be used by those users requiring to have InterProScan graphic output formats for publications and other uses.
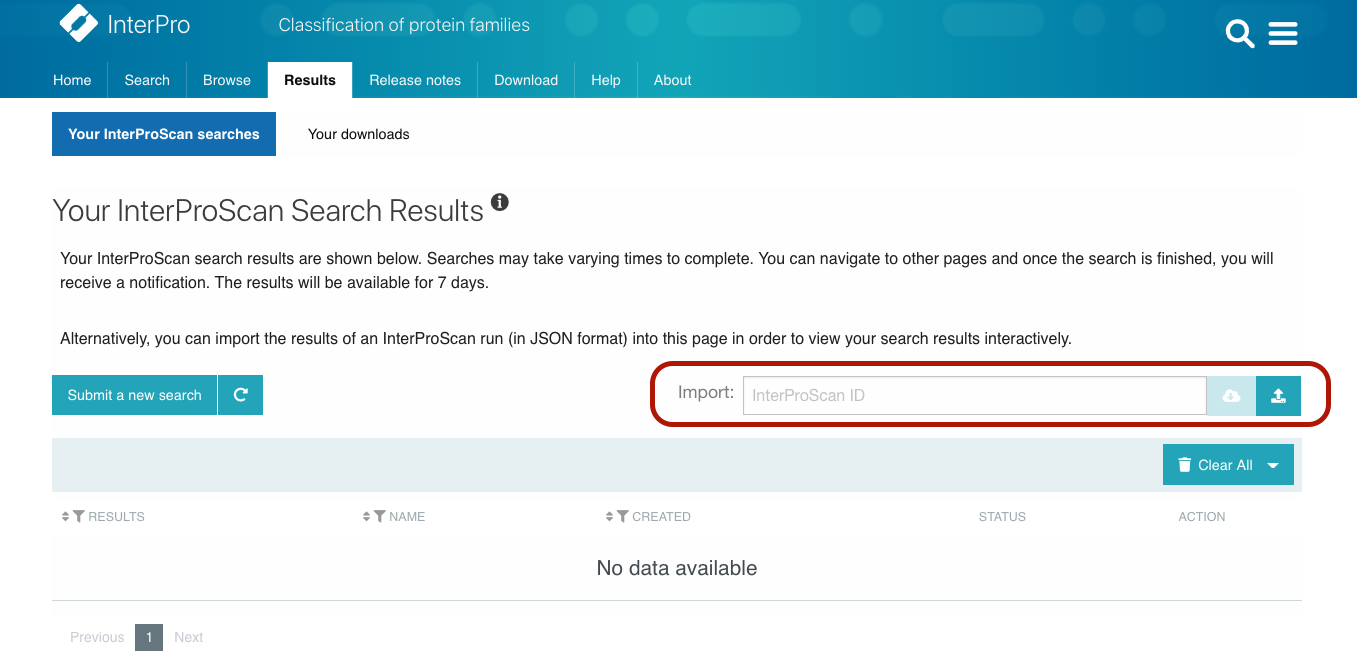 Figure 3. Import InterProScan output files in InterPro.
Figure 3. Import InterProScan output files in InterPro.
The HTML and SVG output formats in InterProScan are now outdated if you compare to the graphical view of the protein page on the website. As such, in the second quarter of 2021, InterProScan will stop rendering HTML and SVG output formats.
Other improvements
Member database logos
We have introduced the display of the member databases logos throughout the website instead of the previous database SVG images, as shown in Figure 4.
 Figure 4. Member databases logos in the InterPro homepage.
Figure 4. Member databases logos in the InterPro homepage.
Member database Signature tab
The representative model that defines a member database signature can be visualized as a logo, using Skylign. This is displayed under the Signature tab in Member database entry pages. This feature was previously only available for the Pfam database, this has been propagated and is now also available for PANTHER, PIRSF, SFLD and TIGRFAMs signatures.
Browser notifications
When performing a sequence search or waiting for data to be ready to download, you can continue to navigate through the website or other browser tabs or applications and you will get a pop-up notification when the jobs or downloads have been completed (this requires the browser notifications to be allowed).
Export functionality
The Genome3D data corresponding to an InterPro Entry can be exported in JSON or TSV formats.
The results of a text search can now be exported in TSV format alongside the JSON format previously available.
In both cases the exported files will include all the results and not only the ones currently displayed (i.e. not limited to the default 20 items).
