The Beauty Of Domain Architecture And Evolution
focusGene duplication serves as a major force in evolution. Duplicated genes can become a playground for evolution, and duplicates can split functions of the ancestral protein (subfuntionalization), or gain new functions (neofunctionalization). Sometimes they are mutated beyond rescue and become pseudogenes. Here we would like to demonstrate how InterPro can help researchers to navigate a particular group of duplicated proteins and their taxonomic distribution using their domain architecture. This can help us to understand how the protein evolved and diversified and help us to reach certain taxonomic groups that might be otherwise overlooked.
Example 1: HAP1 and TRAK1/2
HAP1 (Huntingtin-associated protein 1), TRAK1 and TRAK2 are paralogues found in humans, eutherian mammals and other animals. These proteins share an N-terminal domain, known as the HAP1_N domain (IPR006933), and also have common functions such as intracellular receptor trafficking. However, the Milton domain (IPR022154, we named it “Trafficking kinesin-binding protein, C-terminal” in InterPro) localised at the C terminus of TRAK1/2 is missing in HAP1, and TRAK1/2 has different functions in mitochondria transportation.
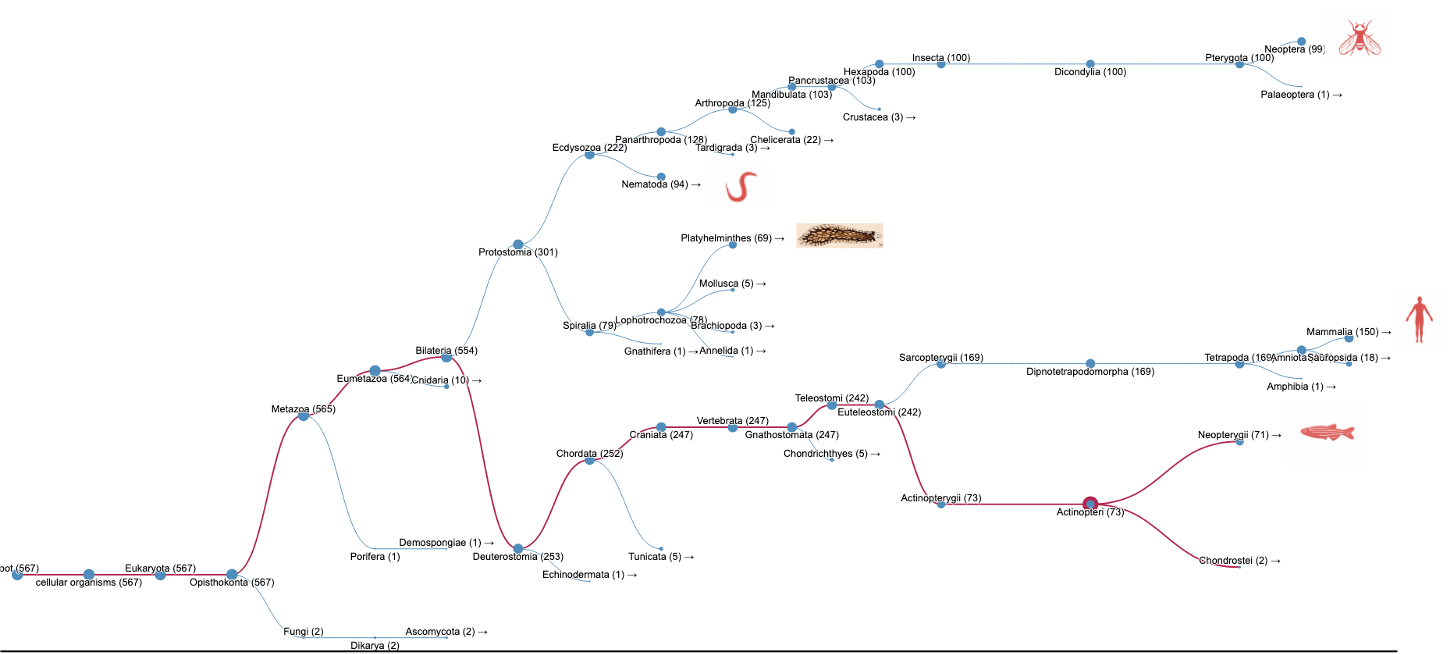
When we searched InterPro using our Search by domain architecture option with the HAP1_N domain (IPR006933) but without the Milton domain (IPR022154), a list of HAP1-like proteins was found (Fig.1A, list [see here]). The analysis of the taxonomic distribution of this group of proteins showed that HAP1-like proteins are mainly found in insects, worms, mammals and fish (Fig.1B). This result is similar to a study published by Keatin et al. [1]. However, we noticed that HAP1-like proteins (around 100 proteins) are found in some species under the Neopterans Infraclass of Insecta. Several species, such as Aphis glycines, Bombyx mori, Apis mellifera and Drosophila sechellia, have one HAP1-like protein and one Milton-like protein. The HAP1-like proteins (such as B4HY25) in these species appear to be small proteins (about 100aa). There are no reports for these group of small HAP1-like proteins and their function so far. Nevertheless, they all contain the HAP1_N domain and share protein sequence similarities with each other.
Figure 1A.
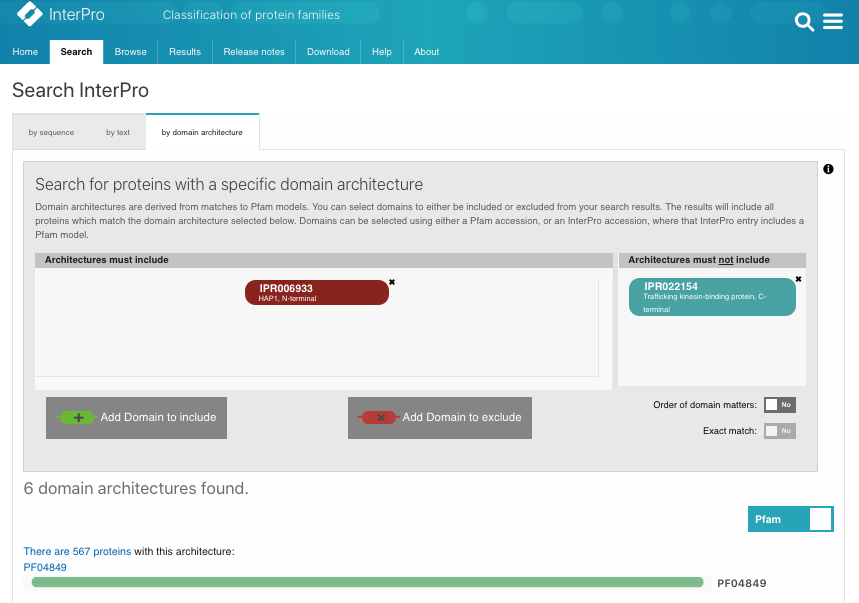
Figure 1B.
Example 2: TPX2 and WDL
TPX2 (targeting Protein for Xklp2) from Xenopus laevis is a microtubule-associated protein that targets a plus end-directed motor (Xklp2) to the minus ends of microtubules in the mitotic spindle. It plays an important role in spindle assembly during mitosis [2]. TPX2 has a highly conserved TPX2 domain (IPR027330, we named it “TPX2 central domain” in InterPro), an Aurora-A binding domain and a TPX2_C domain. However, in plants besides the TPX2 homologue (with three domains), several proteins were found with only the TPX2_C domain. This group of TPX2_C domain only proteins in plants has been named as WDL (or WVD2-like) proteins, as the first identified member is known as WVD2. These WDL proteins bind to microtubules; however, they have lost the function of targeting Xklp2 and were found to play roles in plant development and responses to environment cues [3, 4].
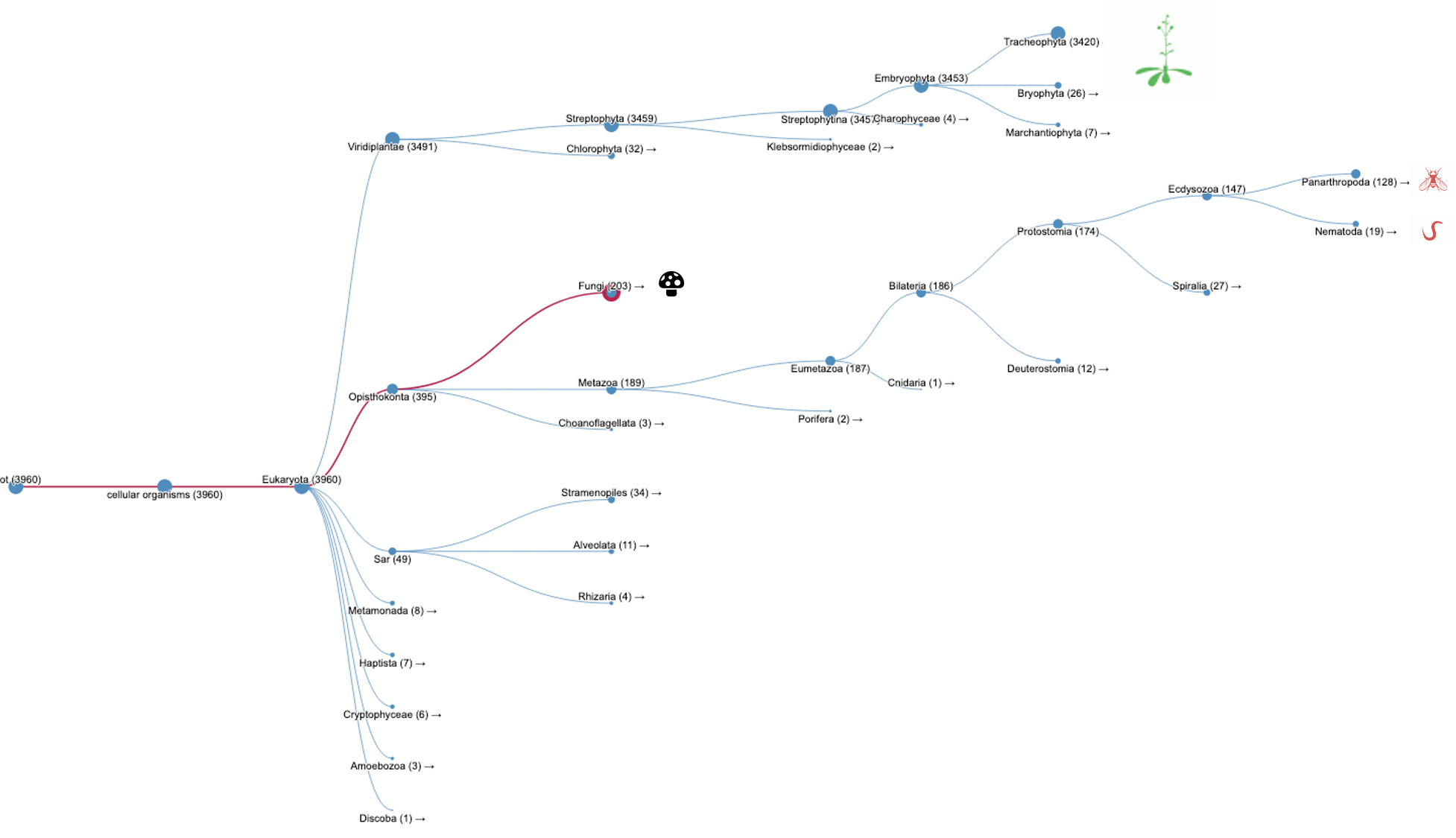
Again, we can search InterPro with the TPX2_C domain (IPR027329) but without the TPX2 central domain (IPR027330). A list of WDL proteins can be found (Fig.2B, list [see here]) and then analysed. It shows that most of the WDL proteins are found in plants (especially flowering plants). Some WDL proteins can be found in insects (such as Ssp1/Mei-38 in Drosophila) and other organisms. From the literature, it has been shown that Drosophila Ssp1/Mei-38 has very little contribution to cell division [5]. As for plants, several WDL paralogues (WDL1-7) have been identified in Arabidopsis. WVD2 and WDL1 have been shown to modulate helical organ growth and anisotropic cell expansion [3], while WDL3 is involved in light-regulated hypocotyl elongation [6].
Figure 2A.
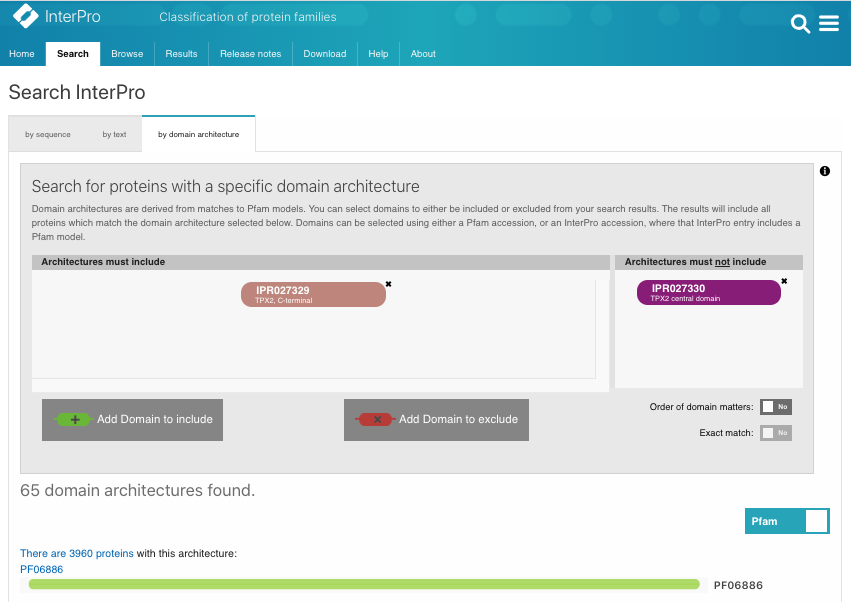
Figure 2B.
It can’t be denied that the vast sea of the NGS (Next-generation sequencing) data and the associated gene predictions can contain errors and noise. Thus care must be taken when trying to decide if the loss or gain of a domain is genuine. Nevertheless, the analysis shown here could serve as a starting point and an effective way to see things from a wider angle.
References
-
Lumsden AL, Young RL, Pezos N, Keating DJ. Huntingtin-associated protein 1: Eutherianadaptation from a TRAK-like protein,conserved gene promoter elements, andlocalization in the human intestine. BMC Evol Biol, 13;16(1):214. 2016. [PMID: 27737633]
-
Wittmann T, Wilm M, Karsenti E, Vernos I. TPX2, A novel xenopus MAP involved in spindle pole organization. J Cell Biol. 149(7):1405-18. 2000.[PMID: 10871281]
-
Yuen CYL, Pearlman RS, Silo-Suh L, Hilson P, Carroll KL, Masson PH. WVD2 and WDL1 modulate helical organ growth and anisotropic cell expansion in Arabidopsis. Plant Physiol, 131(2):493-506. 2003. [PMID:12586874]
-
Sun J, Ma Q, Mao T. Ethylene Regulates the Arabidopsis Microtubule-Associated Protein WAVE-DAMPENED2-LIKE5 in Etiolated Hypocotyl Elongation. Plant Physiol. 169(1):325-37. 2015. [PMID: 26134166]
-
Goshima G. Identification of a TPX2-like microtubule-associated protein in Drosophila. PLoS One.6(11):e28120. 2011. [PMID:26702647]
-
Liu X, Qin T, Ma Q, Sun J, Liu Z, Yuan M, Mao T. Light-regulated hypocotyl elongation involves proteasome-dependent degradation of the microtubule regulatory protein WDL3 in Arabidopsis. Plant Cell. 25(5):1740-55. 2013. [PMID: 23653471]
