Interpro 87.0 New And Updated Features
interproThe latest release of InterPro comes with a number of improvements to the website and API. The list of changes is detailed below.
Website
3D structure viewer
We have updated Molstar to use version 2.x and reduced the amount of text on 3D structure mouse-over, as the text was sometimes cut short when it was too long. It now displays the name of the entry, the chain and residue information, as shown in Figure 1.
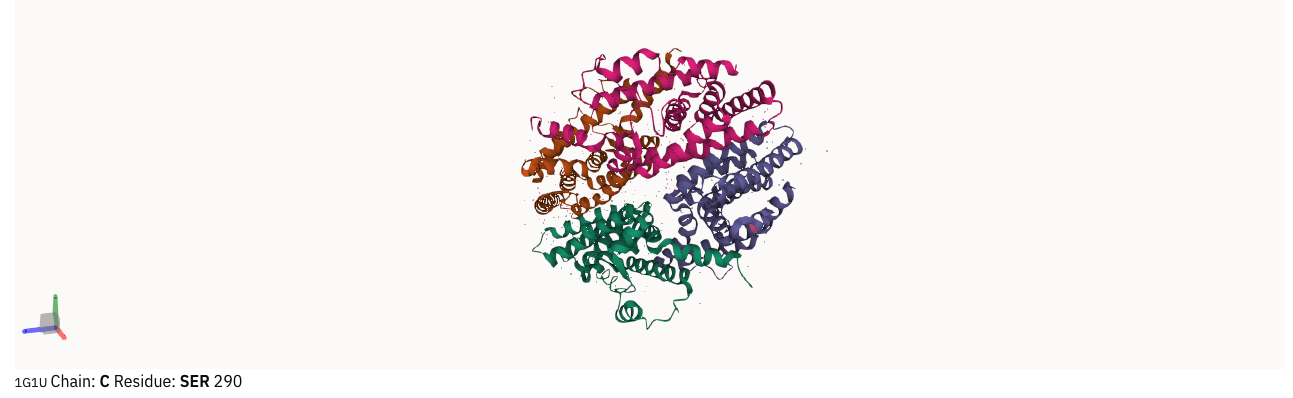 Figure 1. Mouse-over information for 1g1u.
Figure 1. Mouse-over information for 1g1u.
Replacement of trRosetta by RoseTTAFold structure predictions
Following the development of the improved deep learning based modeling method RoseTTAFold by the Baker Lab (Baek, M et al. 2021) and the improved performance of the latter in structure predictions compared to trRosetta (Figure 2), we have made the decision to use this algorithm to predict the structures of Pfam entries.
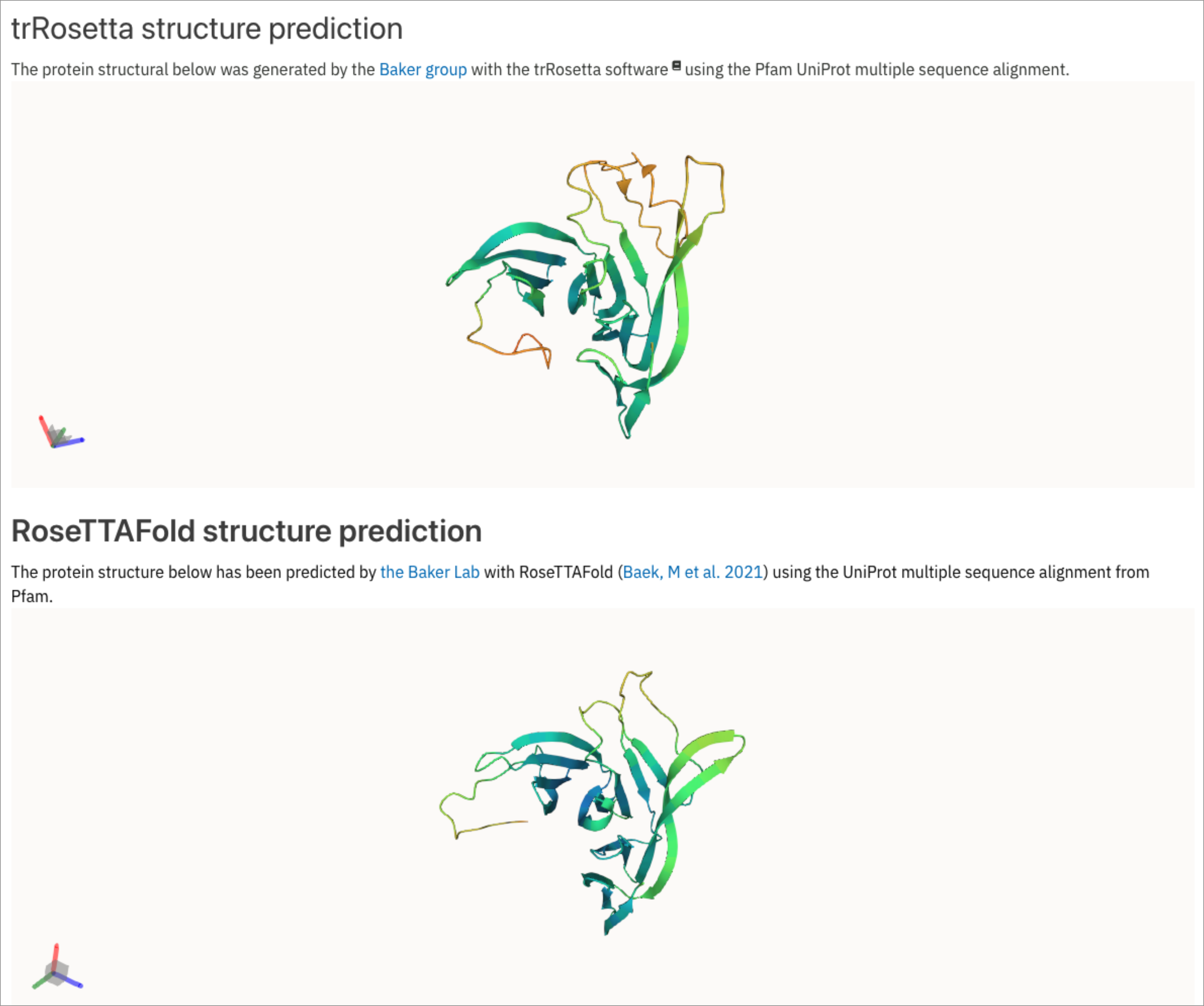 Figure 2. trRosetta vs RosettaFold structure prediction for PF14761.
Figure 2. trRosetta vs RosettaFold structure prediction for PF14761.
RoseTTAFold added to InterPro entry pages
The RoseTTAFold tab has been added to InterPro entry pages, it is displayed when the entry contains a Pfam entry with a RoseTTAFold structure prediction (Figure 3). It provides more visibility to predicted structures at the entry level and will allow an easier comparison with AlphaFold predictions when available for the same entry.
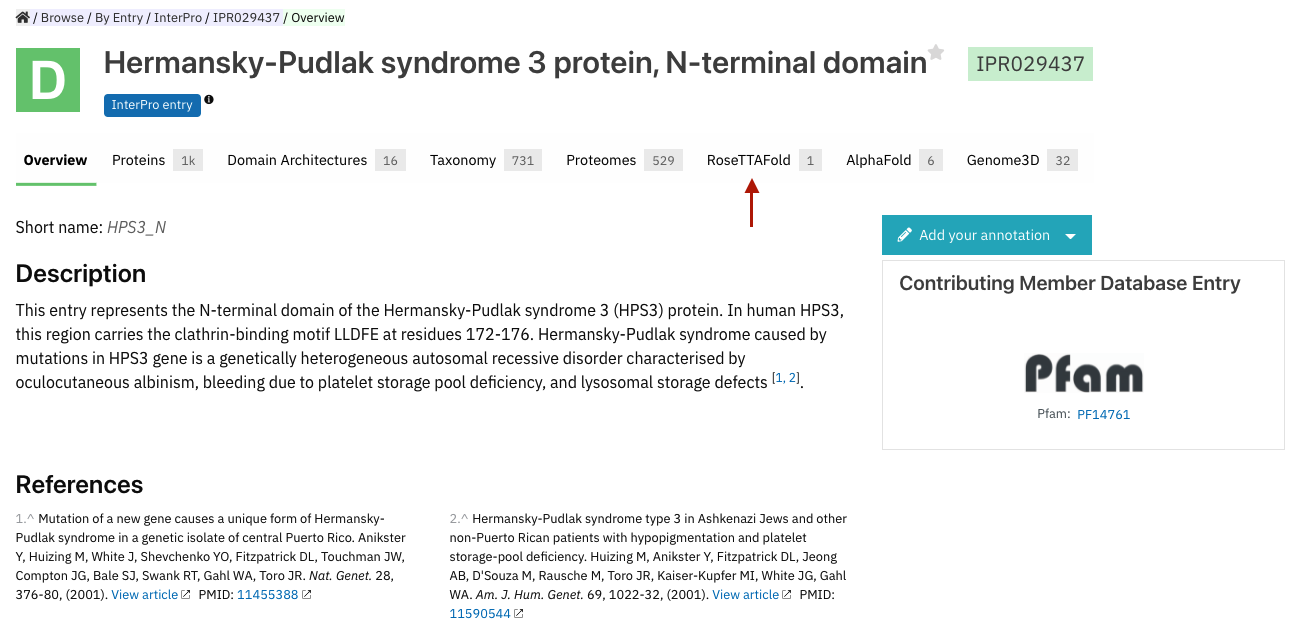 Figure 3. RoseTTAFold tab addition for the InterPro entry IPR029437.
Figure 3. RoseTTAFold tab addition for the InterPro entry IPR029437.
Display structure model matches for Protein pages
The display of the list of AlphaFold structure models in InterPro Entry pages with more than 20 predictions has been changed to a paginated table.
New filter in Browse feature
When using the Browse feature to search for proteins, it is now possible to filter by proteins with/without matches to InterPro entries (Figure 4), in addition to the UniProt curation, taxonomy and Sequence status filters.
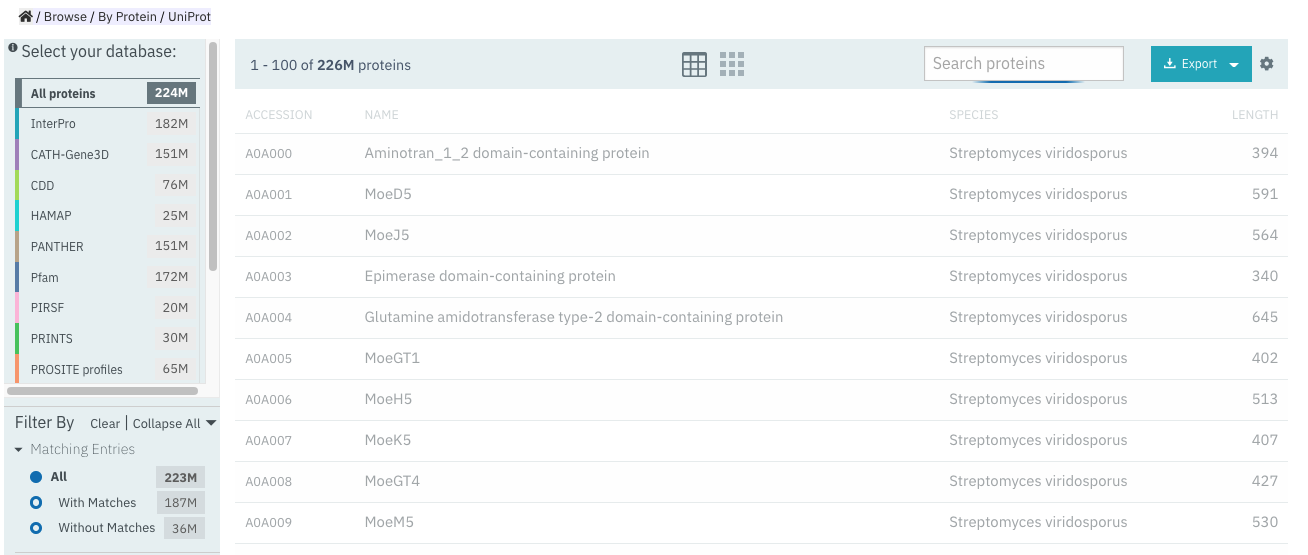 Figure 4. Matching entries filter for proteins in the Browse feature.
Figure 4. Matching entries filter for proteins in the Browse feature.
Upcoming changes
In the next InterPro release (88.0), we are going to introduce greek letters in InterPro entries descriptions. Please contact us if you need help updating your pipeline to support greek letters.
API
As you may be aware, since the redesign of the InterPro website, we have put in place an API to facilitate data programmatic access, accessible through the following link: https://www.ebi.ac.uk:443/interpro/api/. If you are not familiar with the InterPro API yet, you can visit https://www.ebi.ac.uk/interpro/result/download/#/entry/InterPro/, on this page you can apply multiple filters to obtain the data you are looking for, which will help you create the right API URL. On this page we also generate code snippets you can copy and use into your own code.
API documentation update
You may or may not be aware that we have documented the API to help our users get more familiar with the way it works, this is available on GitHub. We have made minor changes to the API documentation for both the user and the developer side.
Added Filter for proteins with Structural Model
When using the InterPro API to get the list of proteins matching an InterPro entry, you can now filter the results to only have proteins with AlphaFold structure predictions by adding the has_model=true option at the end of the url. Example: https://www.ebi.ac.uk/interpro/api/protein/uniprot/entry/InterPro/IPR000001/?has_model=true
Added RoseTTAfold models
Following the replacement of trRosetta by RoseTTAfold models, the API has been updated to access the predicted structures.
