Interpro 88.0 New Features And Improvements
interproThe latest release of InterPro comes with a number of improvements to the website and API. The list of changes is detailed below.
Taxonomy sunburst
InterPro entry pages and Member database entry pages have a Taxonomy subpage. The list of species these entries are matching is based on data from UniProt taxonomy.
The Taxonomy subpage has been reorganised to offer different views, as shown in Figure 1:
- All species table
- Taxonomy tree
- Sunburst
- Key Species table
 Figure 1. Taxonomy subpage view options.
Figure 1. Taxonomy subpage view options.
Sunburst is a new way to visualise taxonomy data in InterPro. The other views were part of the app already, and they have just been reorganised on this page. A sunburst visualisation is a multilayer pie graph that compresses a lot of hierarchical information into a limited space. It shows at a glance the proportions of a variable of interest at different levels of the hierarchy.
The new view in InterPro displays the taxonomy distribution of the proteins matching the entry, from the least specific at the centre to more specific going towards the outside. For example, in Figure 2, the user can infer from the graph that for PF00120, most of the matches are in bacteria, and more specifically in proteobacteria.
Sunburst is now the default view of the subpage. A range of options can be selected to customise the view:
- The segment size can be adjusted based on the number of sequences matching a taxon (default) or by the number of species per taxon.
- The sunburst depth can be adjusted between 2 to 8 rings.
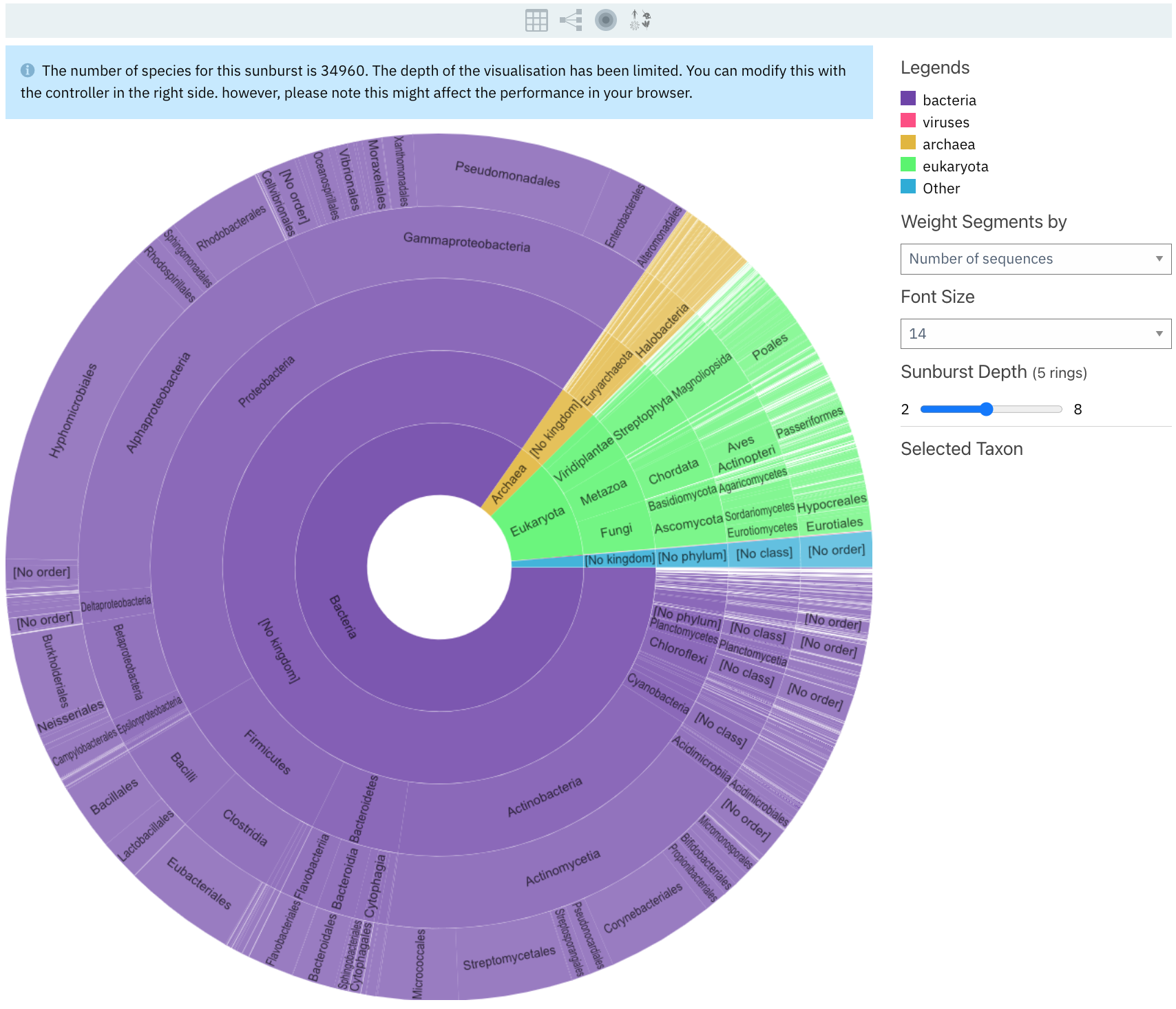 Figure 2. Taxonomy sunburst view for PF00120.
Figure 2. Taxonomy sunburst view for PF00120.
Domain architectures
Domain architectures provide information about the different domain arrangements for the proteins matched by an entry based on Pfam signatures. This information can be found under:
- The Domain architectures subpage of an InterPro entry or member database entry
- The Similar proteins subpage of a protein
- The Results of a search by Domain architecture
Previously, the domains throughout the protein length were displayed next to each other with equivalent sizes, as shown in Figure 3A. In order to display a more biologically correct visualisation, the domain size is now displayed based on the real length of the domain, using a protein of reference. When hovering over a domain, more details are available in a tooltip, including the domain’s position, as shown in Figure 3B.
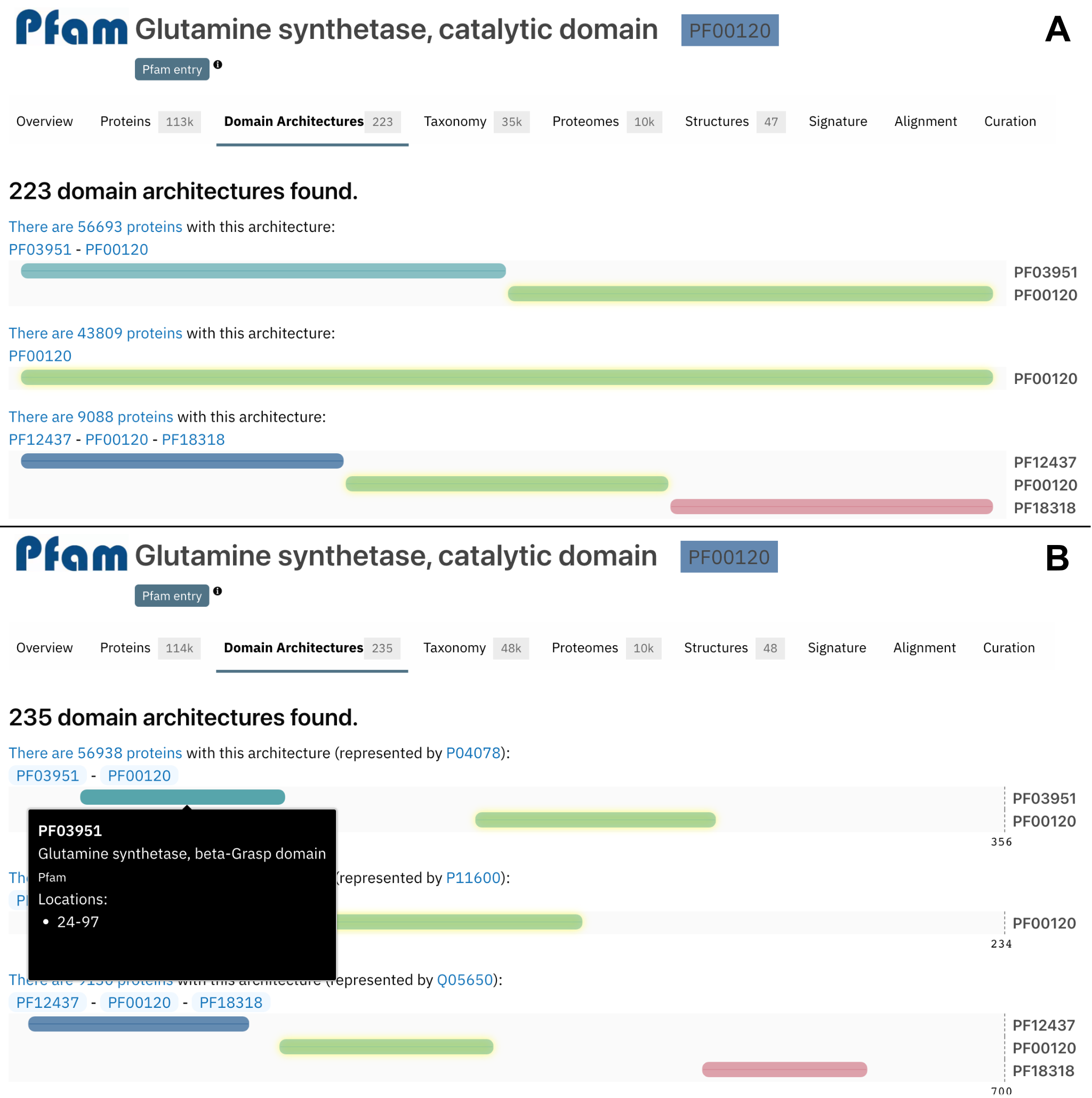 Figure 3. Domain architectures display changes between A) InterPro 87.0 and B) InterPro 88.0 for PF00120.
Figure 3. Domain architectures display changes between A) InterPro 87.0 and B) InterPro 88.0 for PF00120.
API documentation update
The InterPro API documentation consists of General documentation available on GitHub and a Swagger API documentation allowing to apply a range of modifiers to the different API data types to filter the output data. The Swagger API has been updated to take into account the modifiers recently implemented. Additionally, a document containing all the modifiers and link examples has been generated in the git repository.
Other changes
The InterPro website caches the information in the browser to avoid delays when requesting the same resource multiple times. However, each browser has a limit on the size of the resource that can be stored. Therefore, big payloads from our API are not cached. For example, if a user opens the alignment page of a Pfam entry, then goes to the overview of the entry, and then back to the alignments page; it’s likely that the alignment file is over the browser storage limit, which results in having to download the file twice.With the changes included in this release, large payloads are now compressed before using the browser storage. Hopefully the compression will reduce the size of the resource to be under the browser threshold which would allow it to be cached and avoid the multiple downloads. Unfortunately, if the resource is so big that even after compression it still surpasses the limit, it won’t be feasible to save it. We expect this to only affect a very small number of cases.
The message indicating the entry hits several proteins with AlphaFold models is now displayed only if the entry has more than one AlphaFold model, previously it was displayed for all the entries.
A range of dependencies the InterPro website infrastructure relies on have been updated.
