Interpro 91.0 Latest Developments
interproThis release includes a lot of new developments to the InterPro website and additional annotations from different resources.
PANTHER update
PANTHER is a large collection of protein families that have been subdivided into functionally related subfamilies, using human expertise. However, subfamilies vary a lot between releases, which makes the curation checking long and tedious. Following the update of the PANTHER from version 15.0 to 17.0, we have decided to stop integrating PANTHER subfamilies and focus our annotation efforts onto integrating PANTHER families.
For each PANTHER family, the list of subfamilies can be found under the Subfamilies tab in the signature page as shown in Figure 1. In the protein sequence viewer, the tooltip for PANTHER matches now includes information about the particular subfamily matching the current protein (Figure 2).
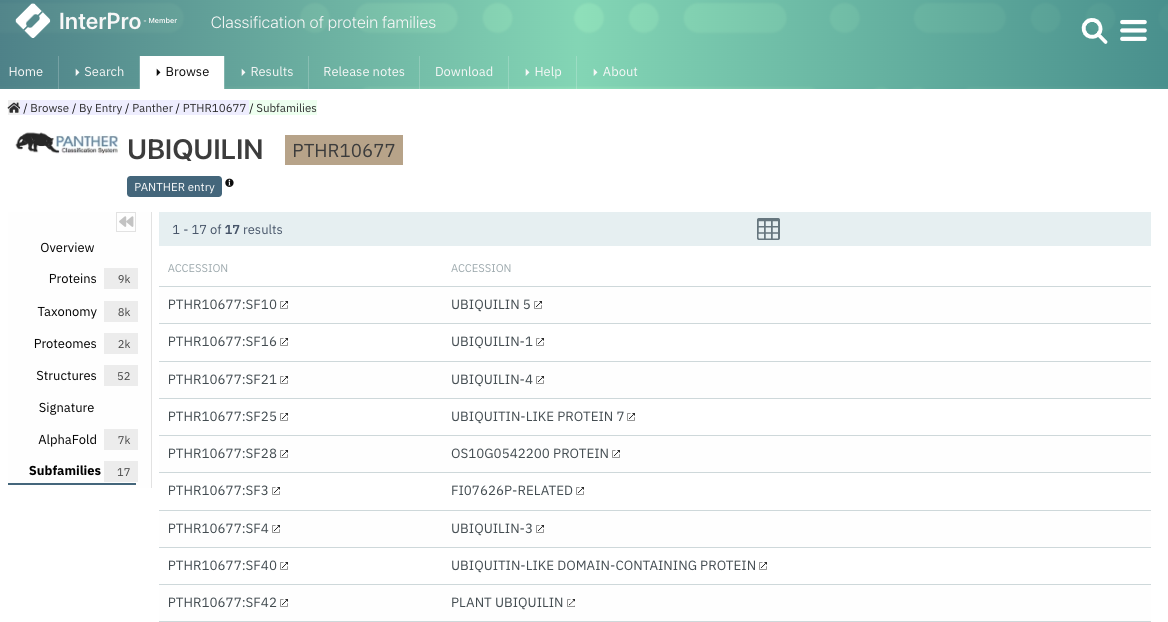 Figure 1. PANTHER Subfamilies tab for PTHR10677.
Figure 1. PANTHER Subfamilies tab for PTHR10677.
Updates to the protein sequence viewer
The style layout of the protein viewer was updated to use CSS grids. This makes better use of the space on the page and fixes a few issues with the space for labels.
As mentioned above, the information about PANTHER subfamilies is displayed in the tooltip of PANTHER family matches (Figure 2).
CATH Funfams
CATH Functional Families (FunFams) is an automatically generated profile HMM database, with FunFams entries segregated by an entropy-based approach that distinguishes different patterns of conserved residues, corresponding to differences in functional determinants. In this release, we have included Funfams to the list of additional annotations provided by InterPro. They are displayed under the Other features section of the protein sequence viewer, as shown in Figure 2.
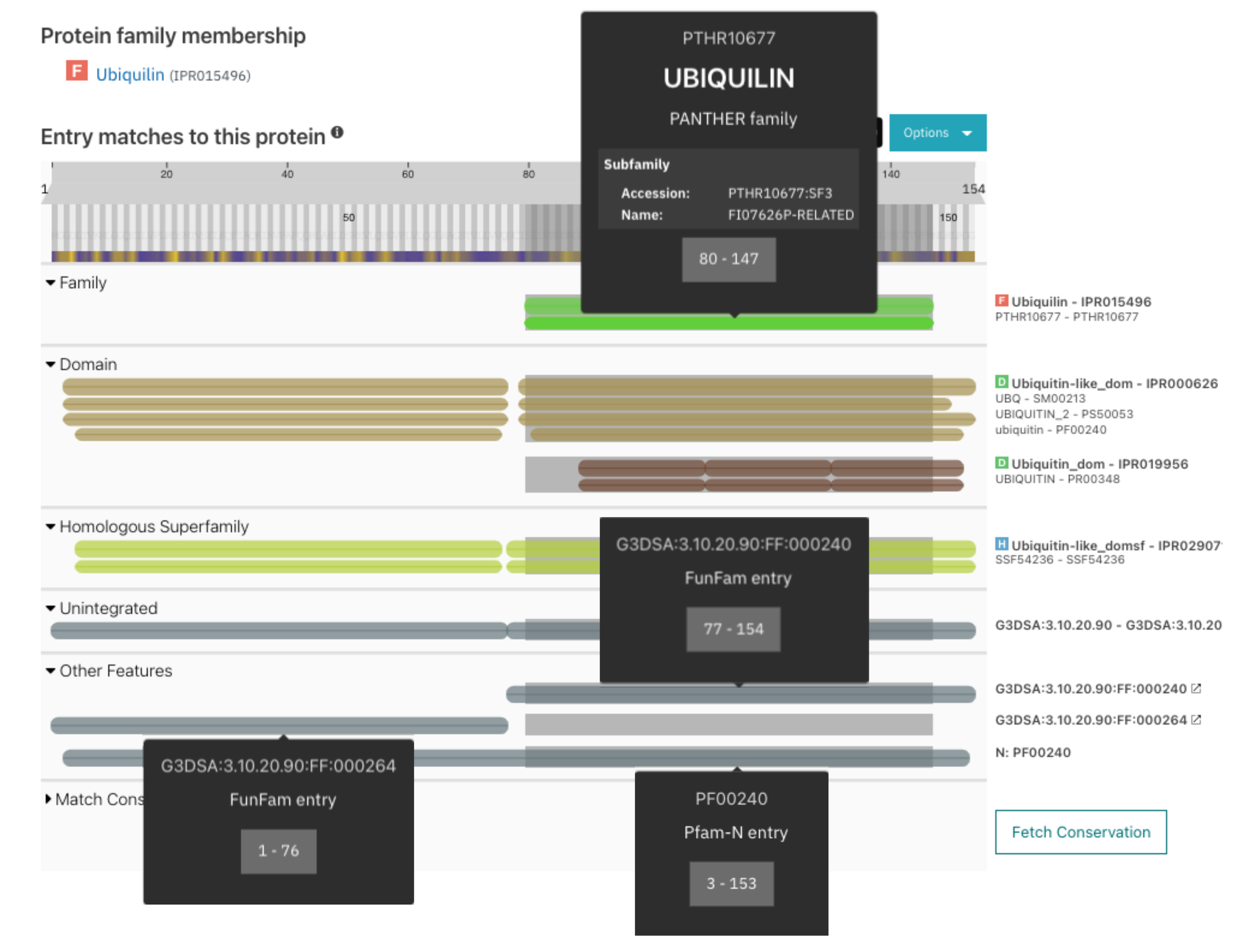 Figure 2. PANTHER subfamily, Funfams and Pfam-N information for O02741.
Figure 2. PANTHER subfamily, Funfams and Pfam-N information for O02741.
Update on AlphaFold data
AlphaFold predicted structures were previously available from protein and InterPro entry pages only. They are now available from the AlphaFold tab for all member database signatures.
The AlphaFold tab on protein pages displays the predicted structure in the Molstar viewer and the InterPro matches in the protein sequence viewer. Hovering over a match in the sequence viewer highlights the corresponding region in the 3D view, as shown in Figure 3.
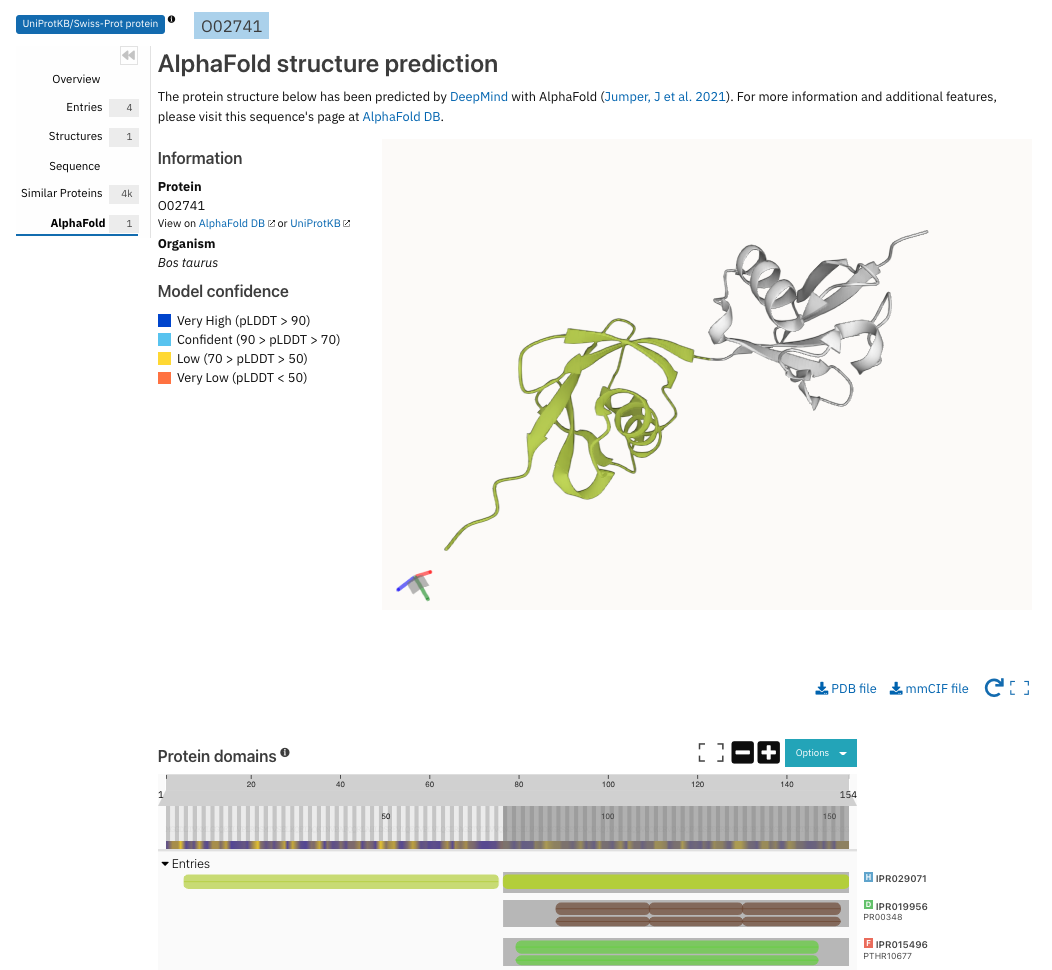 Figure 3. Interaction between the protein sequence viewer and Molstar viewer for AlphaFold structure prediction for O02741.
Figure 3. Interaction between the protein sequence viewer and Molstar viewer for AlphaFold structure prediction for O02741.
Domain architecture
The list of domain architectures are now scaled in reference to the protein with the longest sequence on the page, as shown in Figure 4B.
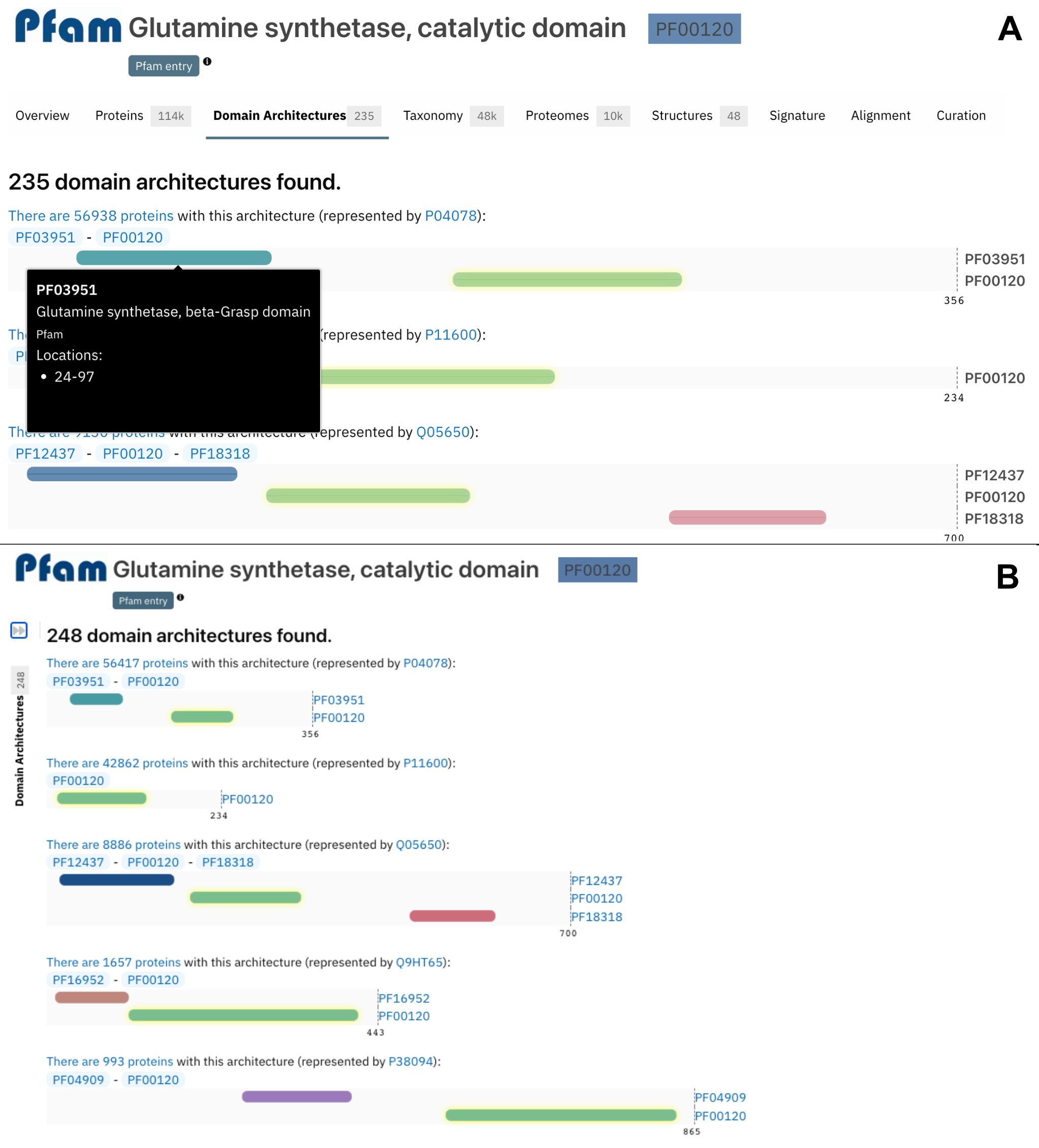 Figure 4. Old (A) and new (B) domain architecture representation for PF00120.
Figure 4. Old (A) and new (B) domain architecture representation for PF00120.
Pfam-related changes
Due to the decommissioning of the Pfam website, we have removed all the links to it.
Different alignments can be visualised from the Alignments tab in Pfam signature pages. The dropdown list allows you to choose the alignment of interest, it now includes the number of sequences for each alignment.
Short name is now included in the list of entries of a clan. Since March 2021, Pfam and the Google Research team led by Dr Lucy Colwell have been collaborating over Pfam-N, a deep learning methodology developed to increase the Pfam coverage of protein sequences. We have now included the Pfam-N annotation for the whole of UniProtKB proteins. These annotations are available in the Other features section of the protein sequence viewer, as shown in Figure 2. More details about these annotations can be found in a Pfam blog post.
Sequence search
The sequence search now supports the submission of multiple protein sequences at the same time, and groups them as a single job in the results, as shown in Figure 6.
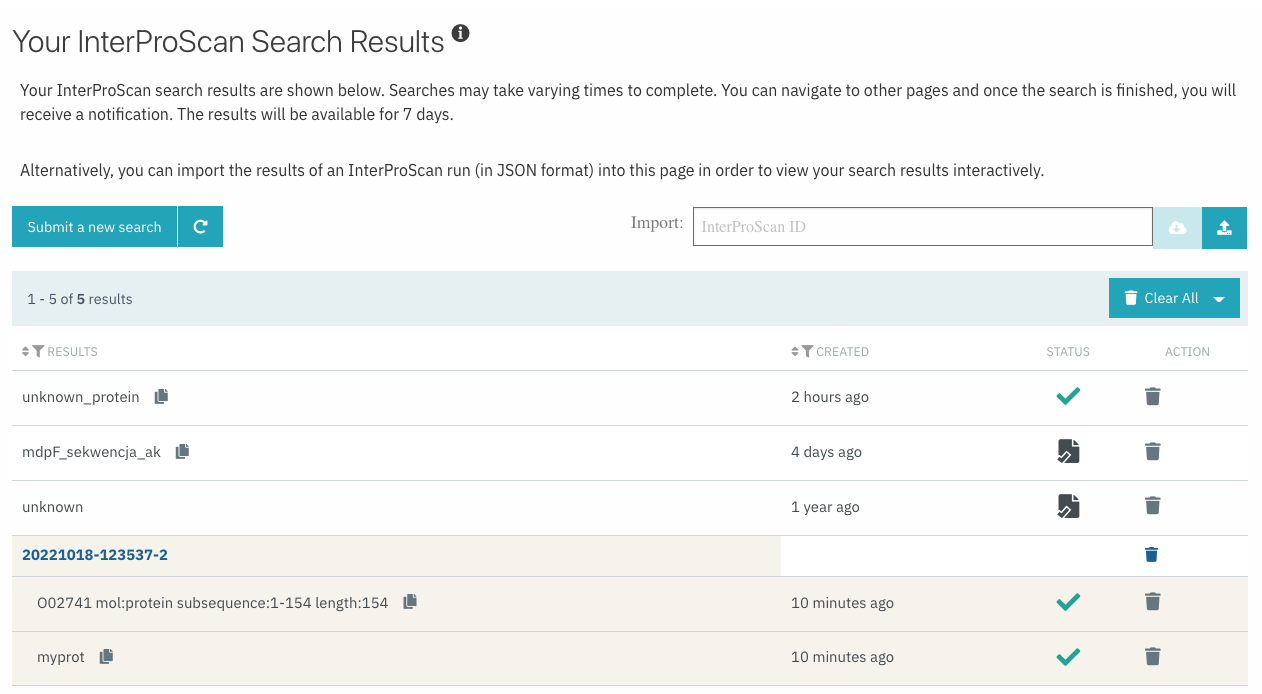 Figure 6. Example of batch search listed in the InterProScan search results table.
Figure 6. Example of batch search listed in the InterProScan search results table.
The Download page allows you to download InterPro, InterProScan, Pfam, Prints and SFLD datasets.
Fixes
The protein sequence viewer in the InterProScan results now allows to display short names, names and accessions.
Issues with the print and snapshot options in the protein sequence viewer layout have been resolved.
