Interpro 92.0: New Features And Updates
interproThis release includes new features and improvements to the InterPro website. The list of changes is detailed below. If you have any feedback or suggestions, please contact us.
Global Core Biodata Resource
InterPro is now a Global Core Biodata Resource. The Global Biodata Coalition (GBC) is a forum for research funders to better coordinate and share approaches for the effective management and expansion of biodata resources around the world. Global Core Biodata Resources include deposition databases that archive and preserve primary research data, as well as knowledge bases that bring together and add value to these data through expert curation and annotation, allowing them to be mined, combined and used to advance research.
Data updates
In this release, we have updated InterPro entries based on UniProt 2022_05 and updated CDD to version 3.20. In total we have created 219 news entries and cover 81.8% of UniProtKB.
New features
New track in the protein viewer
The AlphaFold confidence track is now included in the protein sequence viewer on the protein page and in the AlphaFold subpage, as shown in Figure 1.
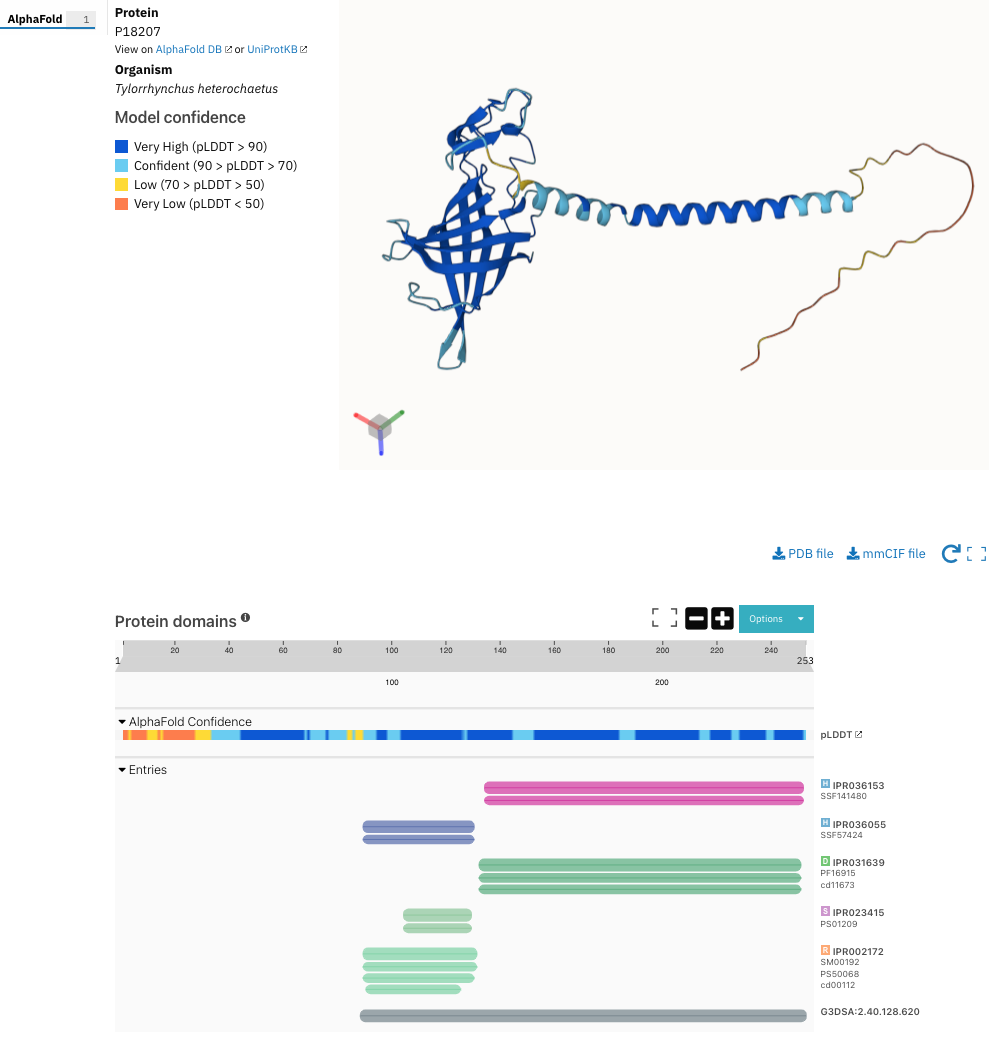 Figure 1. AlphaFold confidence score in the protein sequence viewer for P18207
Figure 1. AlphaFold confidence score in the protein sequence viewer for P18207
PANTHER GO terms
The PANTHER database provides GO terms for families and subfamilies signatures. The GO terms associated with PANTHER subfamilies are now displayed in the PANTHER Subfamilies tab (Figure 2A). Additionally, if a protein has a PANTHER match, the GO terms of the corresponding PANTHER subfamily are displayed at the bottom of the protein page, below the InterPro GO terms, as an expandable section (Figure 2B).
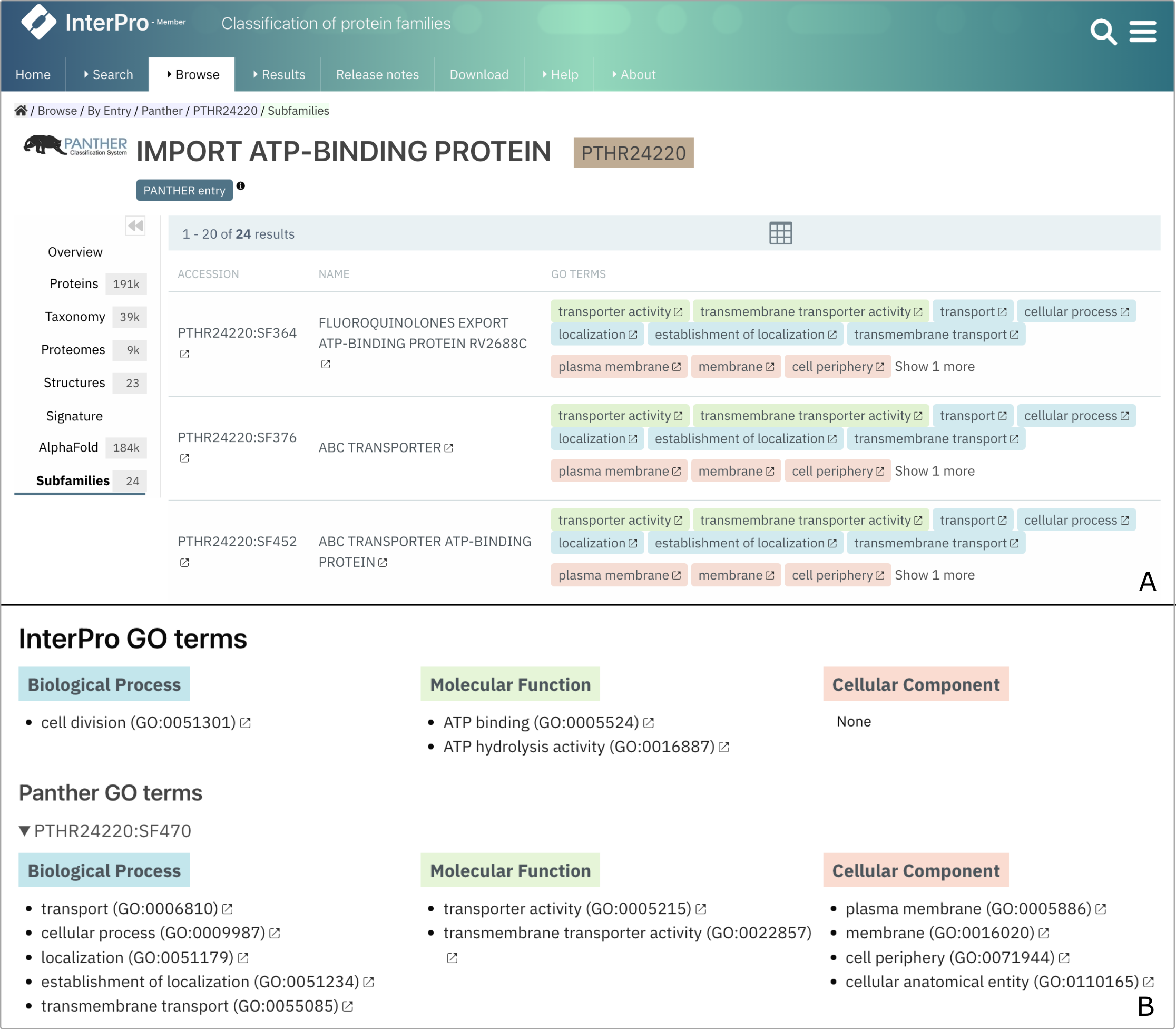 Figure 2. Display of PANTHER GO terms in the PTHR24220 Subfamilies tab (A) and for UniProt P0A9S0 (B)
Figure 2. Display of PANTHER GO terms in the PTHR24220 Subfamilies tab (A) and for UniProt P0A9S0 (B)
Batch actions for InterProScan jobs
As of InterPro 91.0, it is now possible to submit batch sequence searches. In this release, we have added the option to Delete All, Re-run All, and Download All the submitted sequences at once from the InterProScan search result page, as shown in Figure 3.
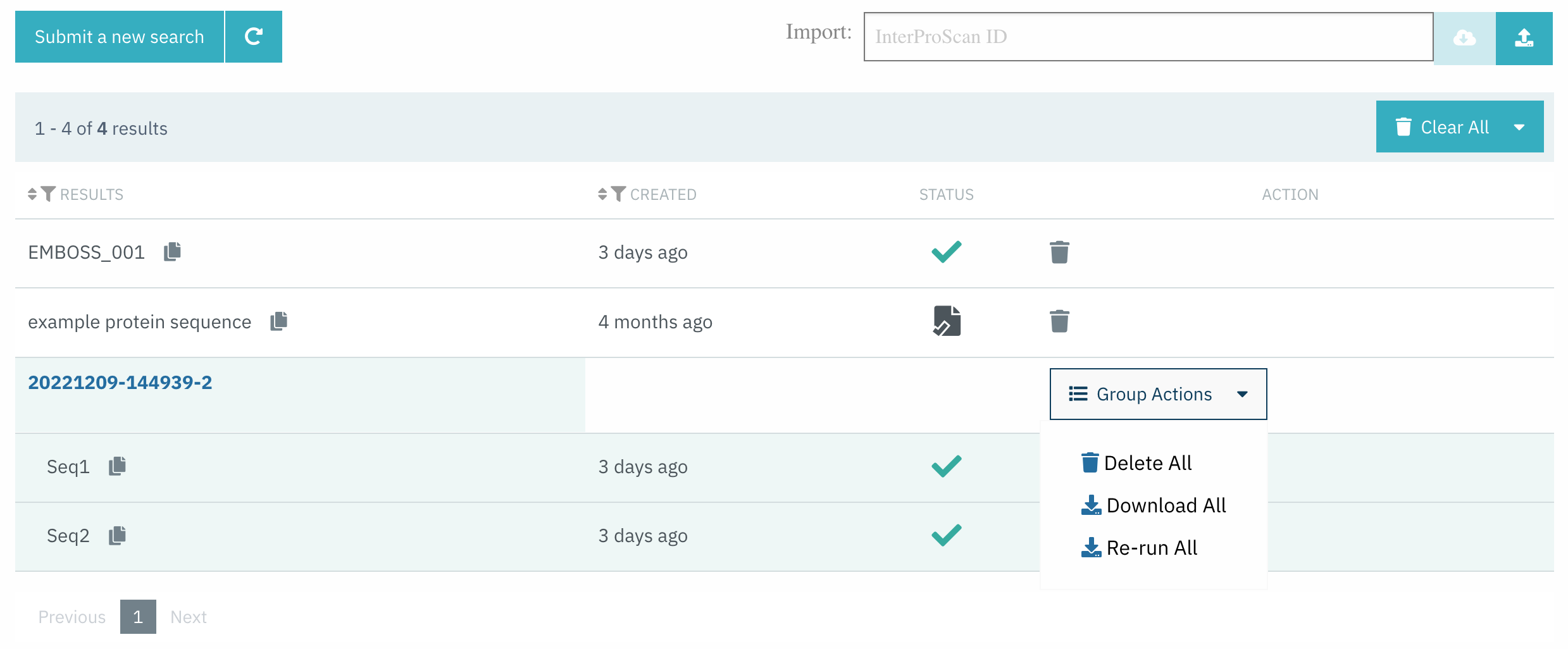 Figure 3. Group actions for InterProScan batch searches
Figure 3. Group actions for InterProScan batch searches
Search by domain architecture results
The search by domain architecture allows to find proteins that contain specific domains. Previously, when searching by domain architecture, the results showed the InterPro or Pfam accessions (Figure 4A). The accessions have now been substituted by their short names (Figure 4B), which are easier to grasp biologically.
 Figure 4. InterPro 91.0 domain architecture representation (A) vs. InterPro 92.0 representation (B) for PF00001-PF00002 search
Figure 4. InterPro 91.0 domain architecture representation (A) vs. InterPro 92.0 representation (B) for PF00001-PF00002 search
Citing InterPro
We have recently published a new InterPro paper: InterPro in 2022. The citation on the website home page has been updated and is now fully displayed by default, whereas it had to be expanded before.
Fixes
Thanks to the feedback of multiple users, we noticed that the downloadable files for Pfam alignments were missing the .gz extension, this has been fixed. Additionally, we discovered that unintegrated Pfam were not rendered in the protein viewer when a Pfam-N entry was also matching the protein; this issue has been resolved.
The tooltips in the protein sequence viewer display the region of the sequence covered by a domain. The region boundaries also serve as a link to the sequence subpage in a given protein, allowing one to view the corresponding residues in the protein sequence. The links were broken in the protein sequence viewer displayed on the structure page; this issue has been fixed.
There was a layout issue in the overlay component covering the set viewer for CDD; the problem has been resolved.
