Interpro 94.0: New Features And Updates
interproThis release includes new features and improvements to the InterPro website. The list of changes is detailed below. If you have any feedback or suggestions, please contact us.
Data updates
In this release we have updated the InterPro entries annotations to support HAMAP database version 2023_01 and replaced TIGRFAMs by NCBIFAMs.
We have also integrated 234 new signatures from the NCBIfam (200), Pfam (1), PANTHER (15), HAMAP (5), CDD (13) databases.
TIGRFAMs replaced by NCBIFAMs
The TIGRFAMs database was transferred in April 2018 to the NCBI and is now part of NCBIFAMs. This release includes an update of the InterPro entries and website features to take into account this change. Additionally, 200 NCBIFAMs have been integrated into InterPro entries.
Migration of the InterPro codebase to TypeScript and Visual framework 2.0
We have started the process to move the InterPro website code base to use TypeScript instead of JavaScript+FlowJS. We will do the migration gradually, migrating the website by sections. To ease the transition, we have added TypeScript support in the bundler, linters, and testing.
We are also going to migrate the codebase from using the EBI visual framework 1.4 (with foundation) to using Visual framework 2.0 (Vanilla CSS). In this release, we have mostly added support for both frameworks, to be able to do this migration progressively.
Upgrading to Nightingale v4 components
In the last 2 years, UniProt and InterPro web developers have been working on a new version of the Nightingale components: v4. The protein sequence viewer, sunburst view of the taxonomy, Pfam alignments and RoseTTAFold model pages in the InterPro website, are now using this new version.
One of the key features allowed by this change is the display of the domain short names on top of the domains (if space allows it) in the protein sequence viewer, as highlighted in Figure 1.
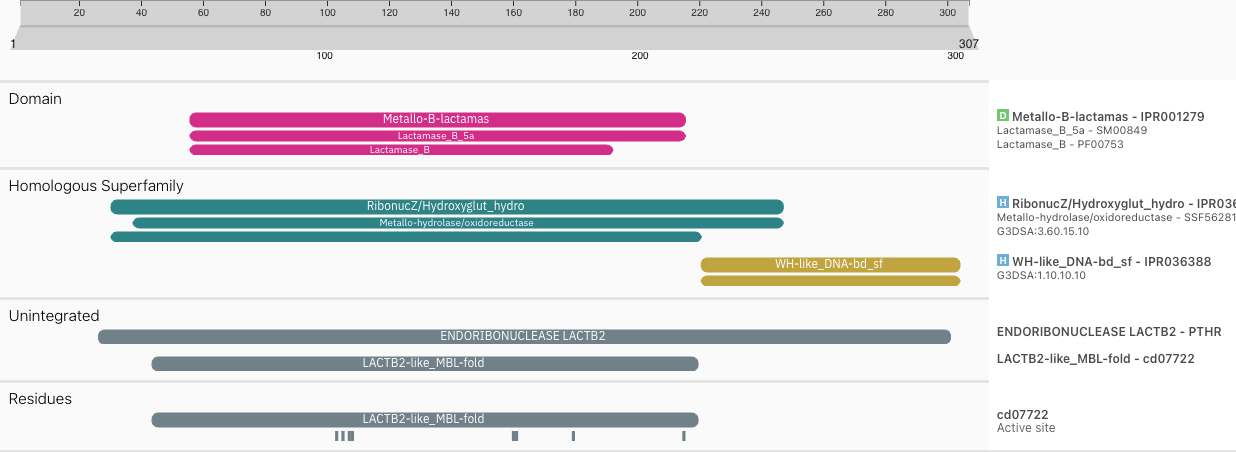 Figure 1. Enhanced protein sequence viewer using Nightingale v4, displaying short names on matches.
Figure 1. Enhanced protein sequence viewer using Nightingale v4, displaying short names on matches.
Breaking Changes
We have drop support for browsers without es6 capabilities. Continuing support of it, was becoming increasingly incompatible with the planned migrations, new features, and in general with our roadmap. Based on our logs, the share of users downloading the legacy version was less than 1%, therefore this change should not affect the big majority of our users. If you however, start having any issues with the new version, please check that your browser is compatible. For a detailed list of which browsers support es6 features, you can go to: https://caniuse.com/es6.
InterPro description provided for integrated signatures without description
Some member databases don’t provide a text describing their signatures. When it is the case, and the signature is integrated in an InterPro entry, the website displays the description associated with the InterPro entry that it is integrated into, an example is shown in Figure 2.
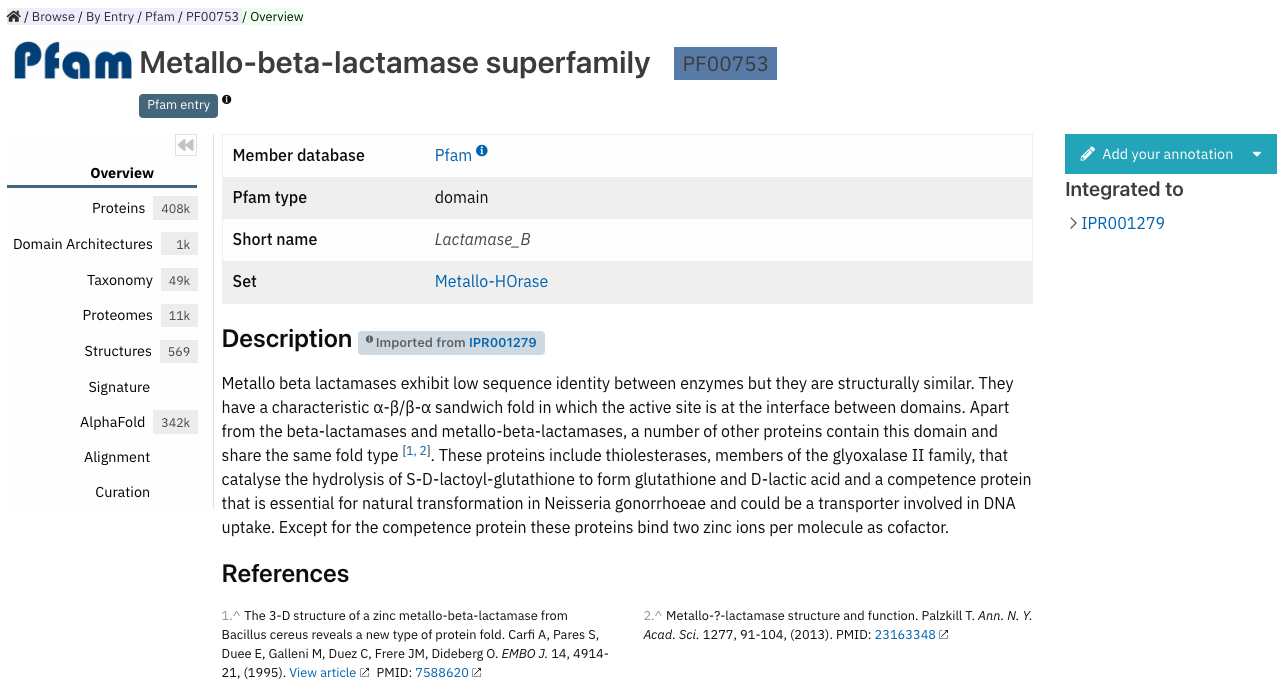 Figure 2. Example of imported InterPro description when the signature doesn’t provide one (PF00753).
Figure 2. Example of imported InterPro description when the signature doesn’t provide one (PF00753).
Updated features for RoseTTAFold 3D viewer
The RosettaFold pages have been updated to offer similar features as the structure and AlphaFold pages, e.g. split screen, disable Picture in Picture.
Fixes
Sequence search result visualisation
When performing a sequence search, the InterProScan results were displaying the matches in a slightly different order than in protein pages. This has been changed and the categories now include “Other features”, “Residues”… as shown in Figure 1.
AlphaFold page
The AlphaFold split screen functionality was displaying an incomplete sequence viewer, this issue has been solved.
The “Short name” in “Label by” option in the protein sequence viewer in the AlphaFold page of a protein was not working, this problem has been solved.
