Interpro 96.0: New Features And Improvements
interproThis release includes new features and improvements to the InterPro website. The list of changes is detailed below. If you have any feedback or suggestions, please contact us.
Content
In this release, we have updated Pfam to version 36.0 and updated InterPro entries accordingly. For more information about the changes in Pfam 36.0, you can have a look at this blog post.
We also have integrated 1,127 new signatures from the Pfam (1,048), CDD (40), CATH-Gene3D (25) and PANTHER (9) databases.
Protein sequence viewer updates
Single domain viewer
A feature found in the Pfam website that was missing in InterPro was to be able to visualise the different domains found in a protein in one line (Figure 1A). We have implemented this feature in the protein sequence viewer on the protein page, as shown in Figure 1B. Please note that this representation is generated automatically using the type of the member databases models, which might differ from the InterPro entries types.
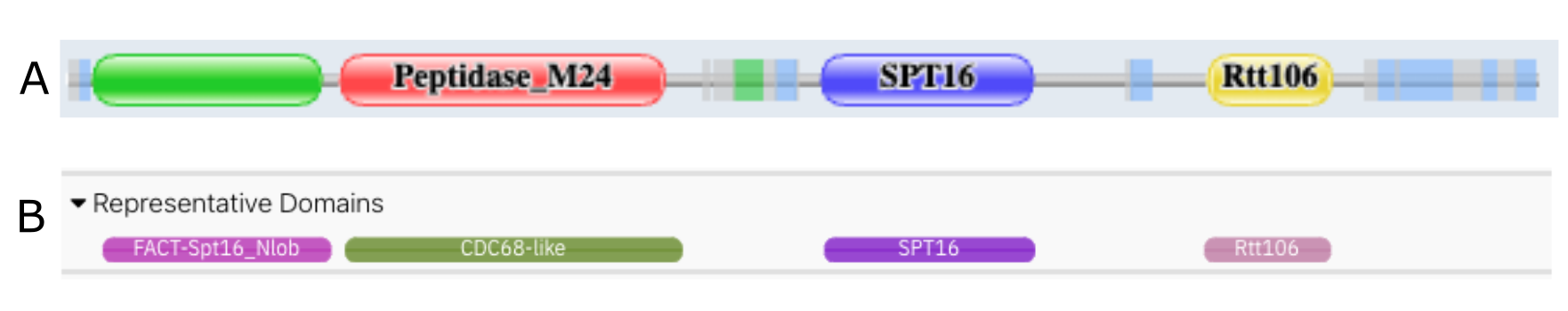 Figure 1. Pfam (A) versus InterPro (B) representative domains view for Q7X923.
Figure 1. Pfam (A) versus InterPro (B) representative domains view for Q7X923.
Tidying up Other features
In the Other features section of the protein sequence viewer, the tracks are now sorted by source database, so all the matches from the same resource are grouped together as shown in Figure 2. Additionally, all the annotations have the source database as part of the label, making it clear where the annotations are coming from.
 Figure 2. Other features section in the protein sequence viewer in InterPro 95.0 (A) versus InterPro 96.0 (B) for Q7X923.
Figure 2. Other features section in the protein sequence viewer in InterPro 95.0 (A) versus InterPro 96.0 (B) for Q7X923.
New features
Addition of AntiFam pages
In InterPro release 88.0 (March 2022), we announced the addition of AntiFam as part of the Other features annotations displayed in the protein sequence viewer. To increase the visibility of AntiFam annotations, we have decided to add AntiFam in the Browse By Member DB (Figure 3A) and create signature pages for each entry (Figure 3B). Additionally, we have moved the AntiFam annotations to the Unintegrated category in the protein sequence viewer (Figure 3C,D). Furthermore, the AntiFam matches are accessible from the Entries tab in the protein page.
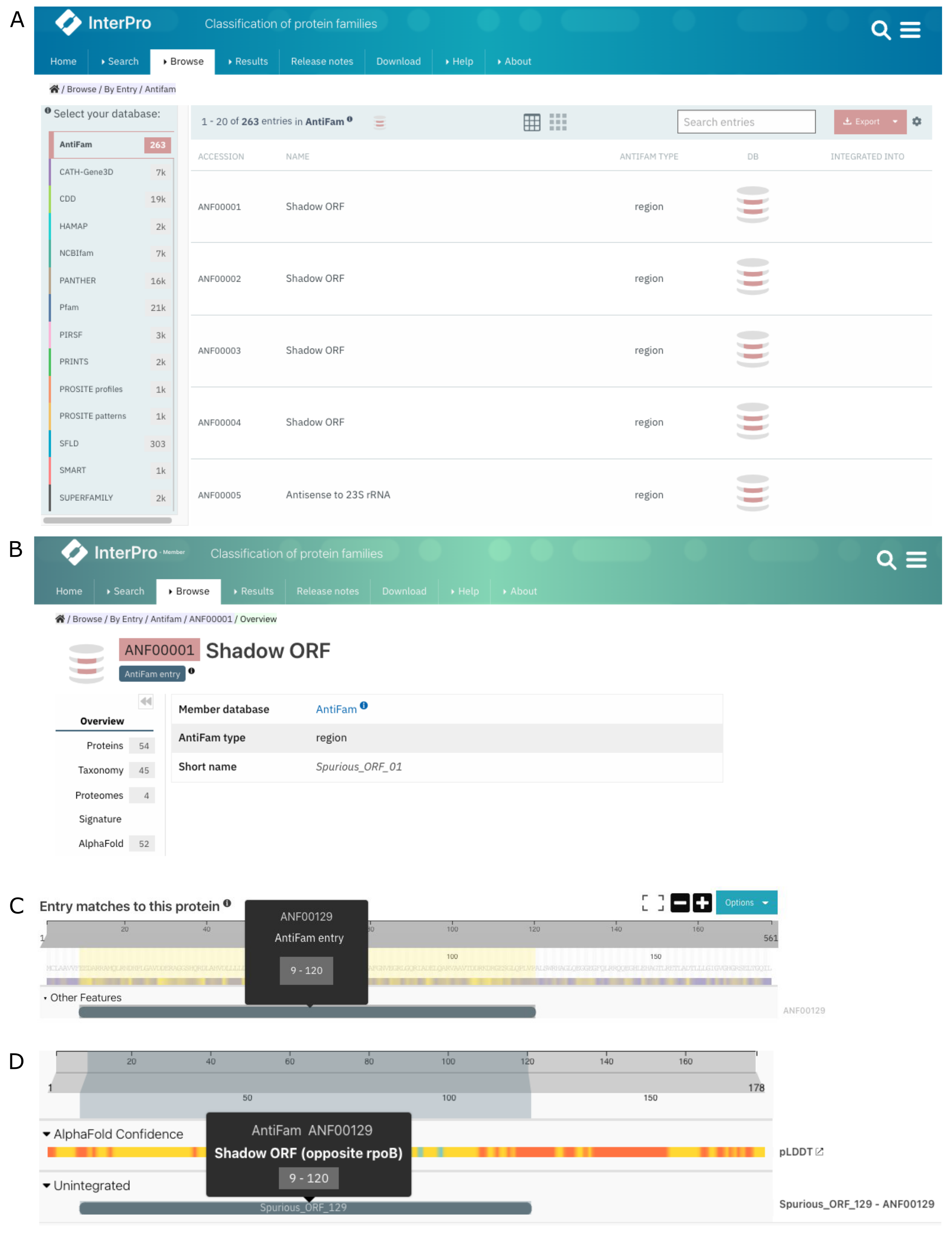 Figure 3. Browse functionality including AntiFam (A). AntiFam signature page (B) AntiFam matches for A0A011PAP6 between InterPro 88.0 (B) and InterPro 96.0 (C).
Figure 3. Browse functionality including AntiFam (A). AntiFam signature page (B) AntiFam matches for A0A011PAP6 between InterPro 88.0 (B) and InterPro 96.0 (C).
Display of structure chains without proteins
PDB structures chains are often associated with UniProt proteins, but not always. On structure pages, we were previously only displaying chains that were associated with proteins. We now display all the chains, whether they are associated with a protein or not.
Additionally, the Entries and Proteins tab in the Structure page and the Structure tab in other pages (e.g. InterPro entry page, Protein page) now display the structure sequence, and a match track for each chain for each structure match, as shown in Figure 4.
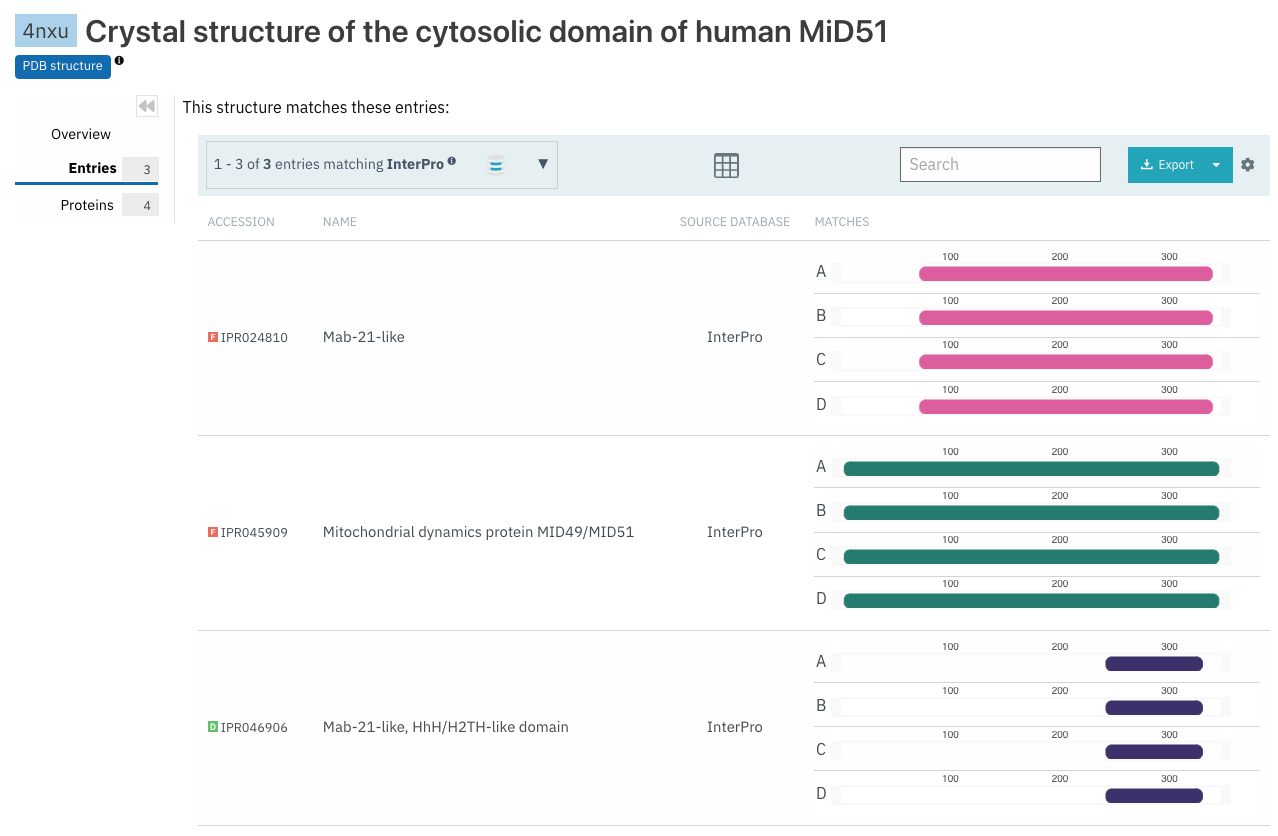 Figure 4. List of InterPro entries and location of the chains matches for the PDB structure 4nxu.
Figure 4. List of InterPro entries and location of the chains matches for the PDB structure 4nxu.
Migration of the InterPro codebase to TypeScript and Visual framework 2.0
As mentioned in a previous blog post, we have started the process to move the InterPro website code base to use TypeScript instead of JavaScript+FlowJS, and we are also migrating the codebase from using the EBI visual framework 1.4 (with foundation) to using Visual framework 2.0 (Vanilla CSS). In this release, we have migrated the following pages and components:
- The Protein page Overview
- The InterPro entry page Overview
- The Signature page Overview
- The Structure page (Overview, Entries, Proteins)
- The Taxonomy, Set and Proteome Overview
