Interpro 97.0: Updates
interproThis release includes new features and improvements to the InterPro website. The list of changes is detailed below. If you have any feedback or suggestions, please contact us.
You can follow us on social media: X: @InterProDB, LinkedIn: InterPro/Pfam for the latest updates.
Post content
- Data updates
- AI-generated descriptions for PANTHER
- Allow search by short names
- Suggesting an annotation update for Pfam signatures
- Replacing “Set” by “Clan” in Pfam-related pages
- Representative domains available via the API
- PANTHER GO terms available in TSV files
Data updates
In this release, we have updated NCBIfam to version 13.0 and PANTHER to version 18.0, and updated InterPro entries accordingly.
We also have integrated 285 new signatures from the NCBIfam (80), Pfam (106), CATH-Gene3D (1), CDD (55) and PANTHER (43) databases.
New features
AI-generated descriptions for PANTHER
Every signature for each member database in InterPro gets its own entry page. These pages contain some basic information such as the signature identifier, the type of entry as defined by the member database (e.g. family, domain or site), and the short name given to the entry by the member database. Some member databases offer a description with details about the family/domain or site function. If the signature doesn’t have a description and is integrated into an InterPro entry, the InterPro description is shown. Nonetheless, there are instances where the member database lacks a description, and the signature is not integrated into an InterPro entry, resulting in the absence of a description. To tackle this, we’ve used AI to summarise information extracted from SwissProt for PANTHER signatures, generating descriptions automatically, an example is shown in Figure 1. It is important to highlight that only a small selection of these descriptions have been reviewed by curators and we advise using them as preliminary sources of information. For more details regarding AI-generated descriptions, you can refer to the InterPro documentation.
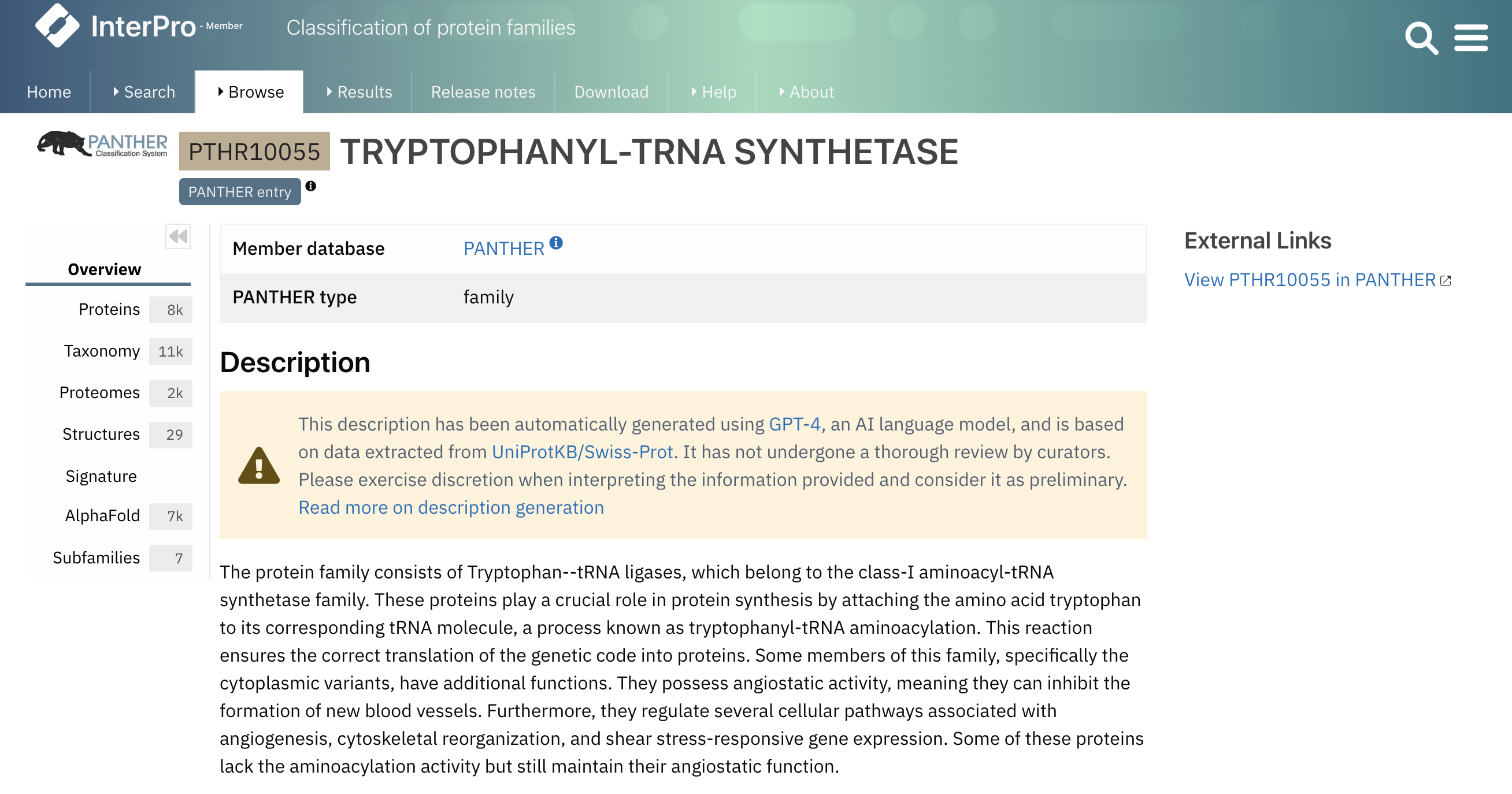 Figure 1. Example of an AI-generated description (PTHR10055).
Figure 1. Example of an AI-generated description (PTHR10055).
Allow search by short names
The InterPro text search functionality allows you to search different types of text:
- InterPro accession (e.g. IPR000562)
- Member database signature accession (e.g. PF00040)
- Protein accession (e.g. P04937) or identifier/short name (e.g. FINC_RAT)
- PDB structure (e.g. 6AR9)
- Gene name (e.g. BRCA2)
- GO terms (e.g. GO:0005911)
- Proteome accession (e.g. UP000000304)
- Taxonomy accession (e.g. 7240)
- Set/Clan identifier (e.g. CL0451)
- Keyword, entry name (e.g. Afadin)
- …
In this release, we have added the possibility to search using the InterPro or member database signature short name (e.g. FN_type2_dom or Neur_chan_memb).
Suggesting an annotation update for Pfam signatures
On Pfam signature pages, the “Add the annotation” button allows users to suggest updates to the Pfam annotation. This was previously pointing to the InterPro help desk, it has now been changed to point to the Pfam helpdesk. Additionally, if you would like to submit data for the creation of a new Pfam entry, you can contact us through this form, providing a multiple sequence alignment and a reference to a publication if available.
Replacing “Set” by “Clan” in Pfam-related pages
Some InterPro member databases (CDD, PIRSF, Pfam) create groups of families that are evolutionary related. Historically, these groups were called clans in the legacy Pfam website. To ease the navigation of Pfam users in the InterPro website, we have changed the naming from Set to Clan in Pfam-related pages.
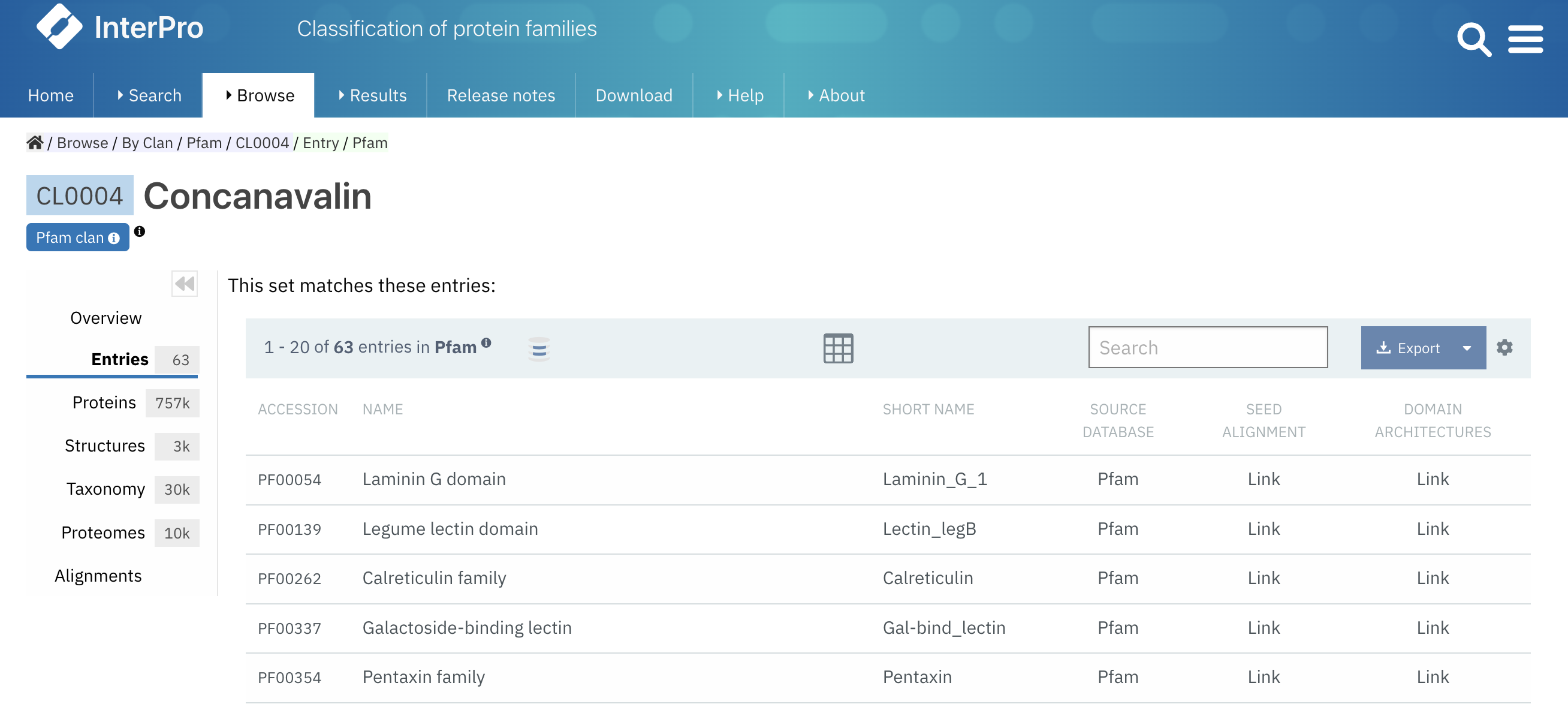 Figure 2. Example of a Pfam clan page (CL0004).
Figure 2. Example of a Pfam clan page (CL0004).
Representative domains available via the API
As part of the last InterPro release, we have implemented a representative domain view in the protein sequence viewer on the protein page, as shown in Figure 3.
 Figure 3. InterPro representative domains view for Q7X923.
Figure 3. InterPro representative domains view for Q7X923.
After multiple user requests, we are pleased to announce that the representative domain information is know available through the InterPro API using the following url structure: www.ebi.ac.uk/interpro/api/entry/all/protein/uniprot/Q7X923 Please notice the newly added key “representative” in the fragments block of the match locations. A value of true indicates that the fragment is considered to be a representative domain. See Figure 4 as an example.
Note: results are paginated (20 results by default); the URL in “next” allows to access the next set of results.
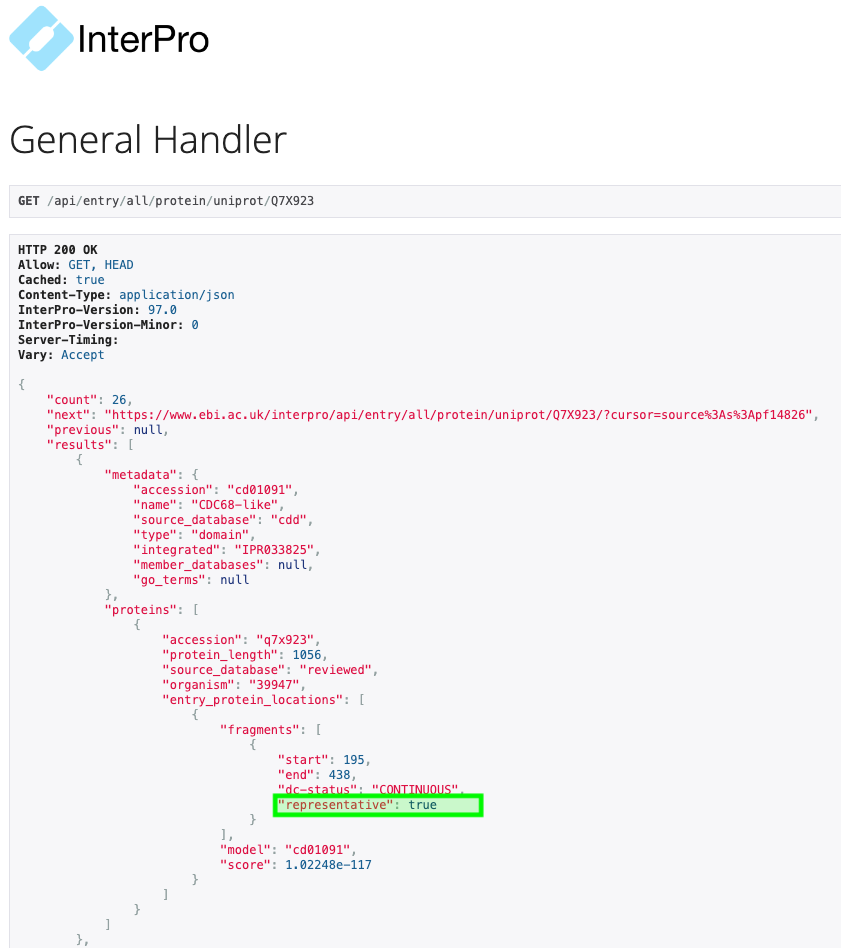 Figure 4. Snapshot of API response for matching entries and signatures on protein Q7X923, highlighting a representeive domain fragment.
Figure 4. Snapshot of API response for matching entries and signatures on protein Q7X923, highlighting a representeive domain fragment.
PANTHER GO terms available in TSV files
When running a sequence search through the InterPro website or through a local installation of the InterProScan software, PANTHER GO terms are reported alongside InterPro GO terms. PANTHER GO terms are now also available when downloading the results in TSV format. An example of TSV file output is shown below.
 Figure 5. Example of InterProScan TSV output.
Figure 5. Example of InterProScan TSV output.
