Interpro 98.0:updates
interproThis release includes new features and improvements to the InterPro website. The list of changes is detailed below. If you have any feedback or suggestions, please contact us.
You can follow us on social media: X: @InterProDB, LinkedIn: InterPro/Pfam for the latest updates.
Post content
- Data updates
- New formats for sequence search batch downloads
- Representative domains in sequence search results
- Consistent categories in the protein sequence viewer
- Search by Domain Architecture
- Contact us page
- Addition of Gene name for protein
- AI-generated descriptions
- Under the hood
Data updates
In this release, we have updated PROSITE patterns, PROSITE profiles and HAMAP to version 2023_05, and updated InterPro entries accordingly. We also have integrated 245 new signatures from the PROSITE profiles (23), NCBIfam (144), Pfam (7), PANTHER (9), HAMAP (1), CDD (61) databases.
New features
New formats for sequence search batch downloads
As you may be aware, it is possible to submit several sequences simultaneously to the InterPro sequence search. When using this feature, the outcomes for all the sequences can be downloaded together. Once the search is finished, the output can be obtained in FASTA, TSV, XML, GFF, and JSON formats for a duration of 7 days through the Group Actions button (Figure 1). Subsequently, the output can be acquired in JSON format.
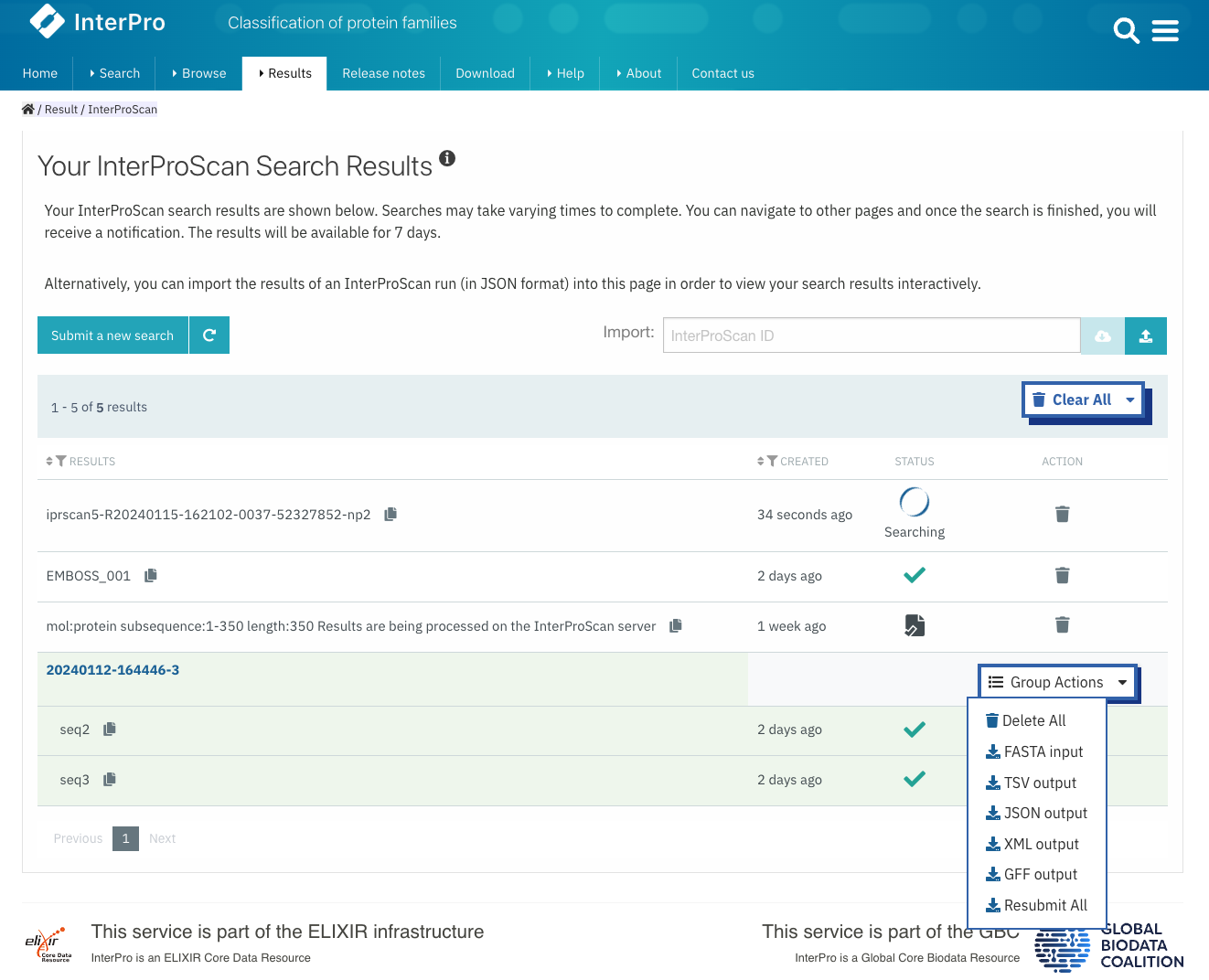 Figure 1. Different download options for a multiple sequences search.
Figure 1. Different download options for a multiple sequences search.
Representative domains in sequence search results
The Representative Domains track enables to easily visualise the different domains composing a protein. This feature was previously accessible in the protein viewer located on the Protein and Structure pages and AlphaFold subpages. Now, it is also accessible in the viewer displaying the outcomes of a sequence search. This is possible thanks to changes in the latest version of InterProScan, which flags the representative domains in its results.
Consistent categories in the protein sequence viewer
We have harmonised the organisation of the different categories shown in the protein sequence viewer for protein, structure and AlphaFold pages. The categories include:
- Representative Domains
- Family
- Domain
- Homologous superfamily
- Repeat
- Site
- Unintegrated
- Other features
- External resources
- Residues
- Other residues
Search by Domain Architecture
The Search by Domain Architecture allows searching for proteins with a particular set of domains. Following user requests, we have added the functionality to download the different domain architectures returned by the search (Figure 2) or found in the Domain Architectures tab of InterPro and Pfam entry pages in TSV and JSON formats using the Export button in the bottom-right corner of Figure 2.
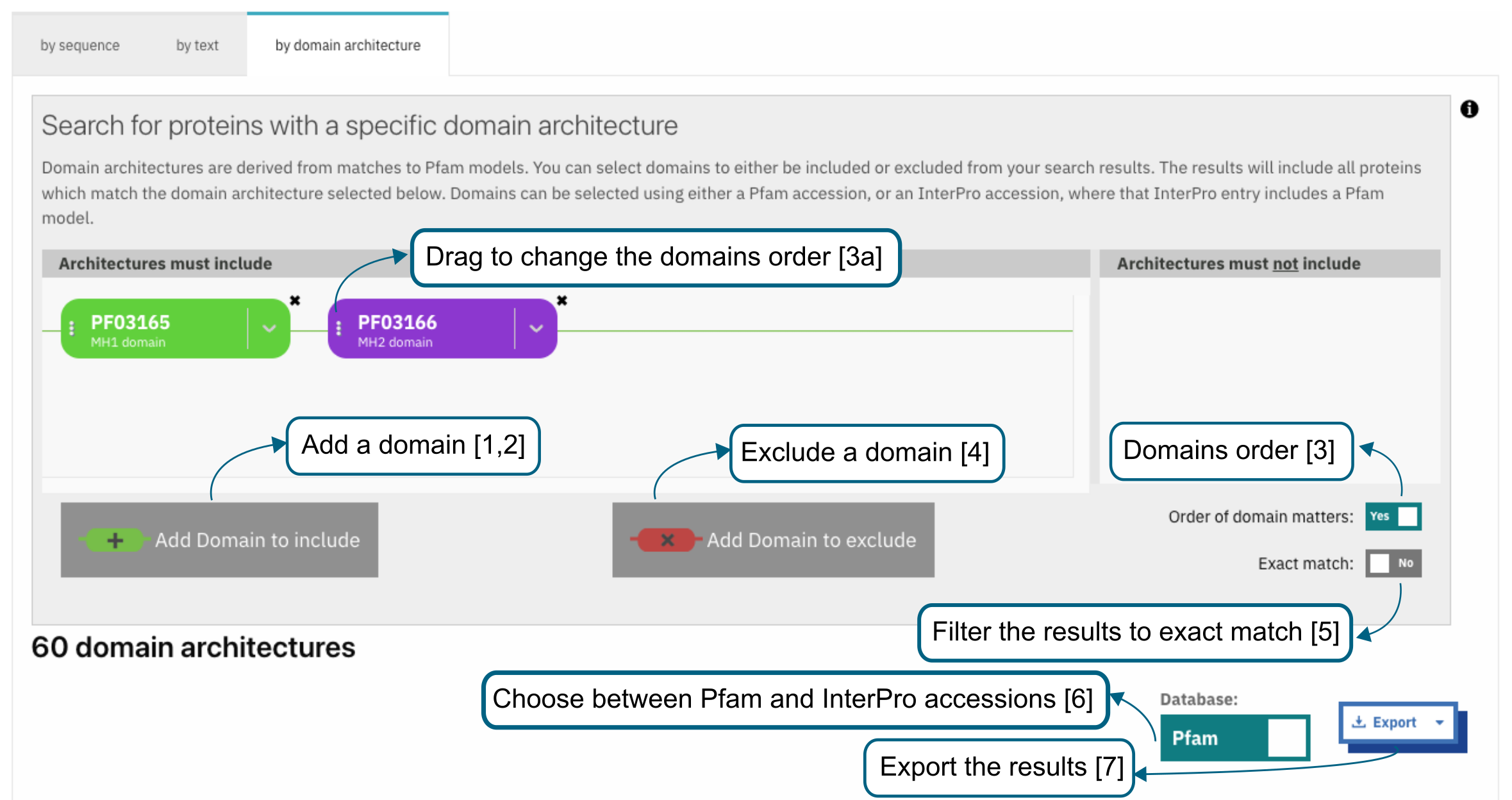 Figure 2. Illustration of how to use the InterPro Search by Domain architecture.
Figure 2. Illustration of how to use the InterPro Search by Domain architecture.
Contact us page
To simplify the process of reaching out to us, we’ve added a Contact us tab to the website header menu, providing direct links to our helpdesks and social media accounts.
Addition of Gene name for protein
In the Overview section of a protein page, we have added information about the gene name encoding the protein. Additionally, the gene name is also shown in tables listing proteins throughout the website (Figure 3).
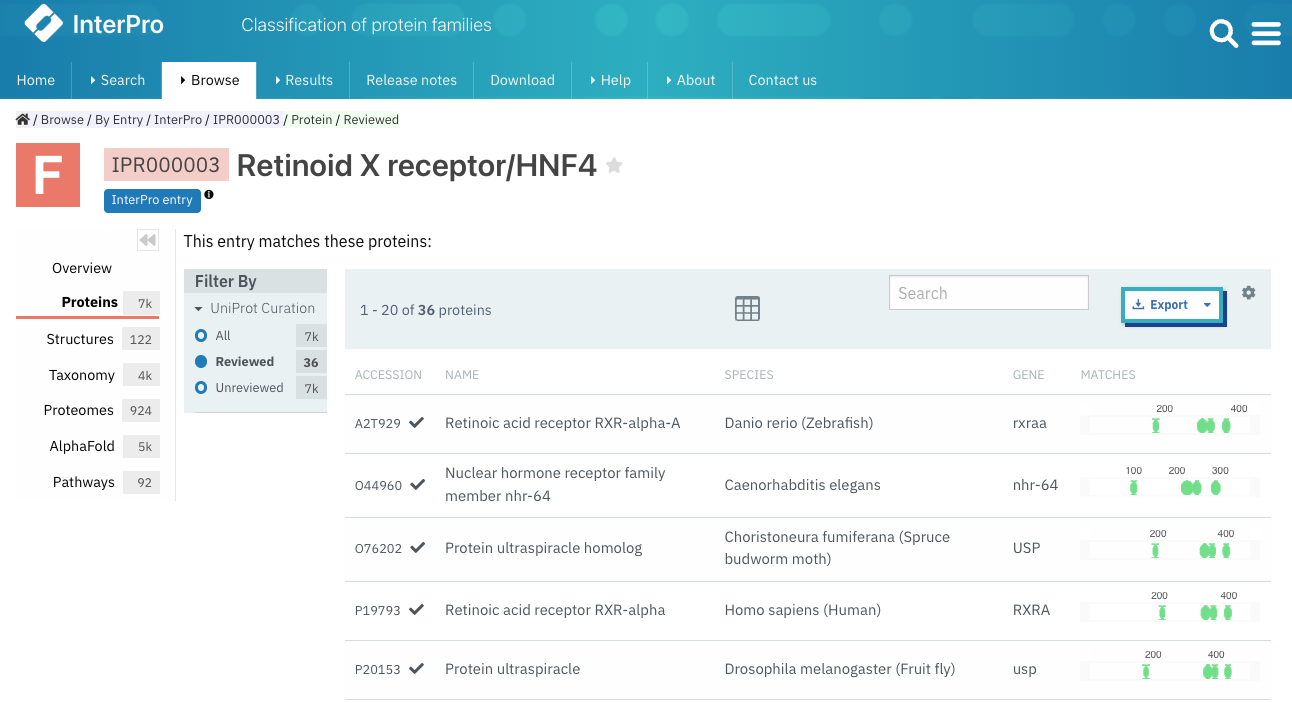 Figure 3. Gene names displayed in the list of proteins for InterPro IPR000003.
Figure 3. Gene names displayed in the list of proteins for InterPro IPR000003.
AI-generated descriptions
We have slightly changed the way we indicate whether a description is generated automatically or not. If at least one of the paragraphs was generated by AI, you may see the following flags:
- AI-generated reviewed: the information provided in the description has been verified by an expert curator.
- AI-generated unreviewed and orange line on the left-hand side: the information provided in the description hasn’t been verified by an expert curator (Figure 4).
- Expert-curated: the description has been written by an expert curator.
For more details regarding AI-generated descriptions, you can refer to the InterPro documentation.
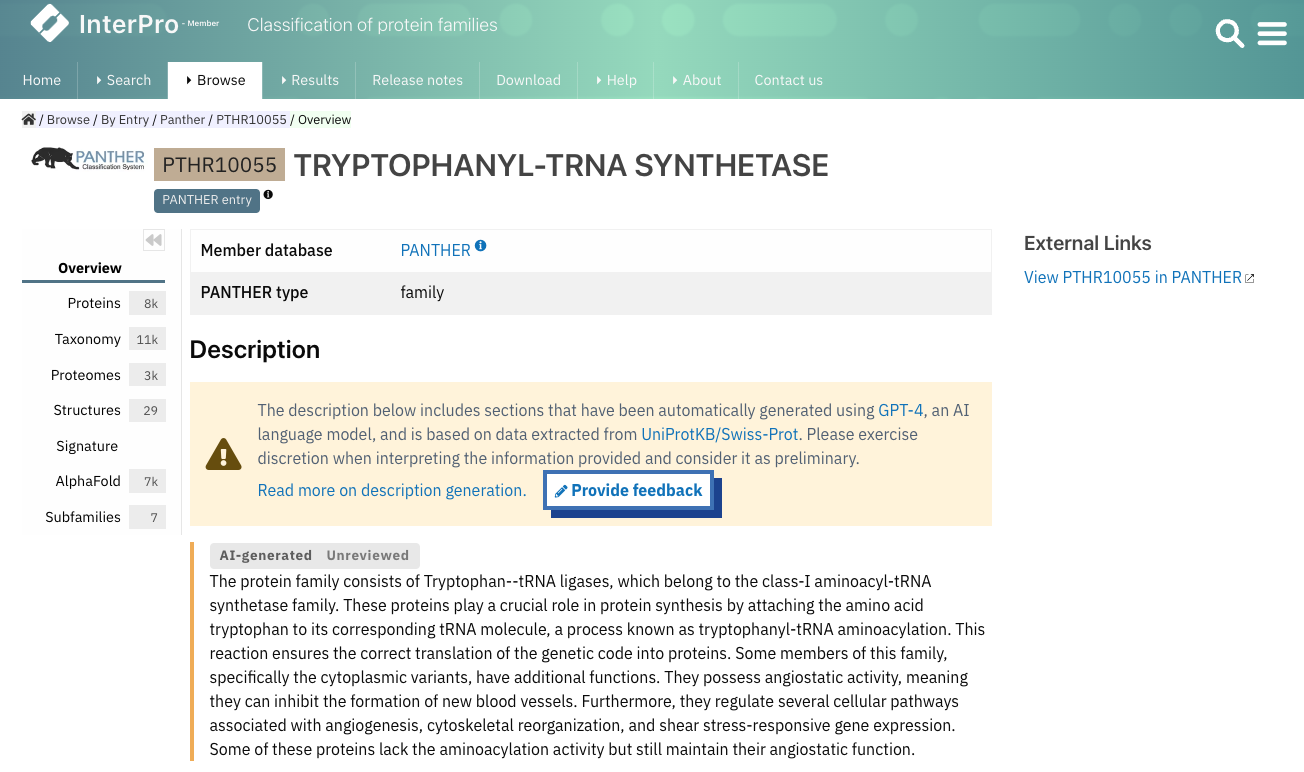 Figure 4. Example of an AI-generated description (PTHR10055).
Figure 4. Example of an AI-generated description (PTHR10055).
Under the hood
We continue our efforts to update the code base of our website. In this release, we upgraded all the dependencies to their latest version and continued the slow but steady migration to TypeScript.
The migration task includes updating the CSS framework. Given that we are doing the migration in gradual steps, it is expected to have a few mismatched design objects, such as buttons on the same page with two different styles. We are working to keep these cases to a minimum. Thank you for your understanding.
