Interpro 99.0: Updates
interproThis release includes new features and improvements to the InterPro website. The list of changes is detailed below. If you have any feedback or suggestions, please contact us.
Post content
- Data updates
- Proteome page
- Link to AlphaFold prediction
- Addition of representative structures
- Citing Pfam
Data updates
In this release, we have updated NCBIfam to version 14.0 and updated InterPro entries accordingly. We also have integrated 3,831 new signatures from the NCBIfam (432), Pfam (1), CATH-Gene3D (64), PANTHER (3,328) and CDD (6) databases.
AI-generated InterPro entries
In this release, there is a big change in the creation of InterPro entries. In the last InterPro release, we introduced AI-generated descriptions for PANTHER signatures. In this release, we went further and introduced AI-generated InterPro entries containing PANTHER, NCBIfam or CATH-Gene3D signatures, which cover more or less the whole protein length (Family).
For these entries, the name, short-name and description have been generated automatically using a Large Language Model. AI-generated content is flagged as such with an AI tag, as shown in Figure 1. For more details regarding AI-generated entries, you can refer to the InterPro documentation.
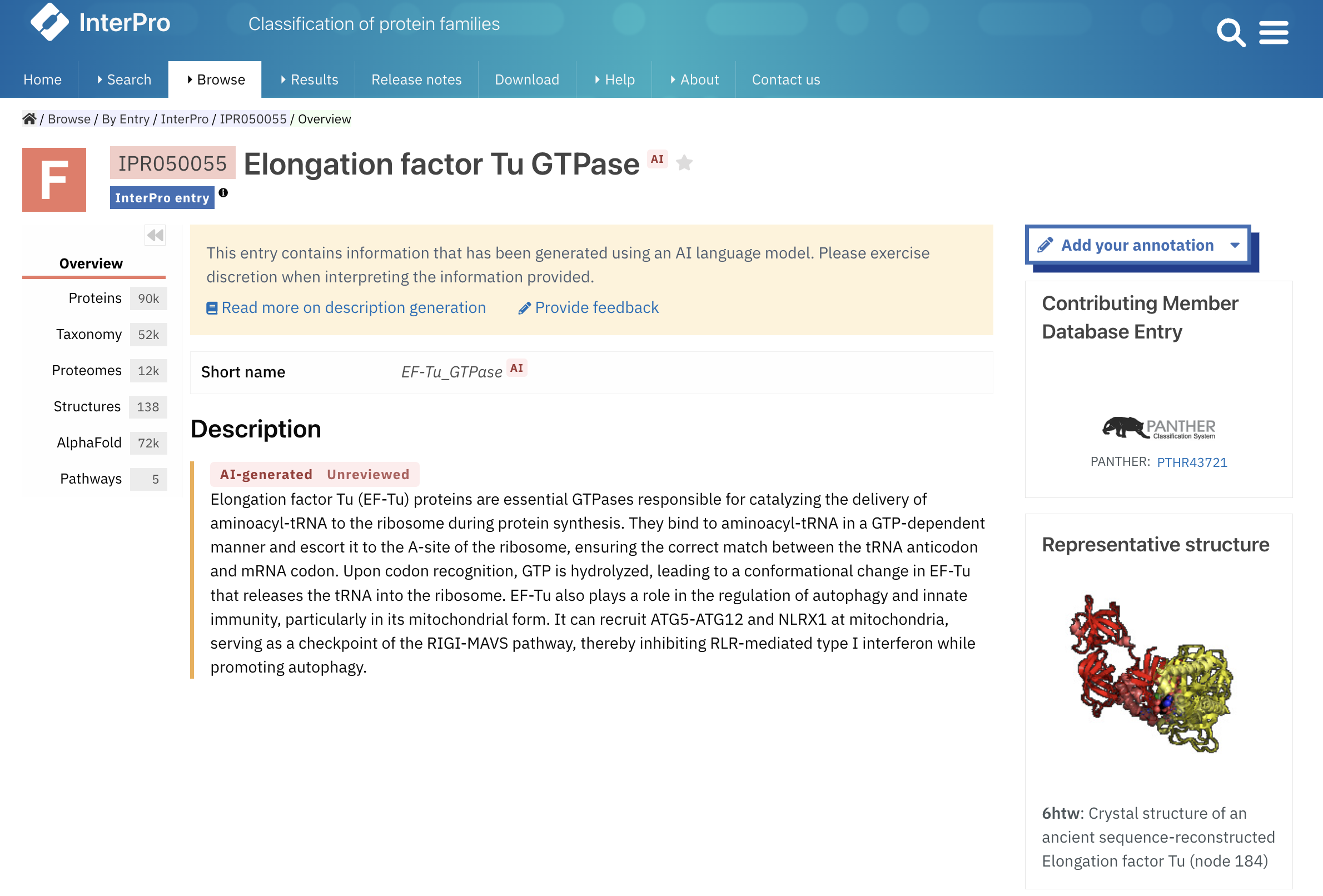 Figure 1. InterPro entry with AI-generated name, short name and description (IPR050055).
Figure 1. InterPro entry with AI-generated name, short name and description (IPR050055).
New features
Proteome page
In the Overview of the proteome page, we now provide a description of the proteome (imported from UniProt) when available and we have added a link to the corresponding Rfam genome page when available to the list of External links.
In the Entries tab of a proteome page, you can now filter the entries by member database e.g. PANTHER, Pfam… using the database selector located in the table header, as shown in Figure 2.
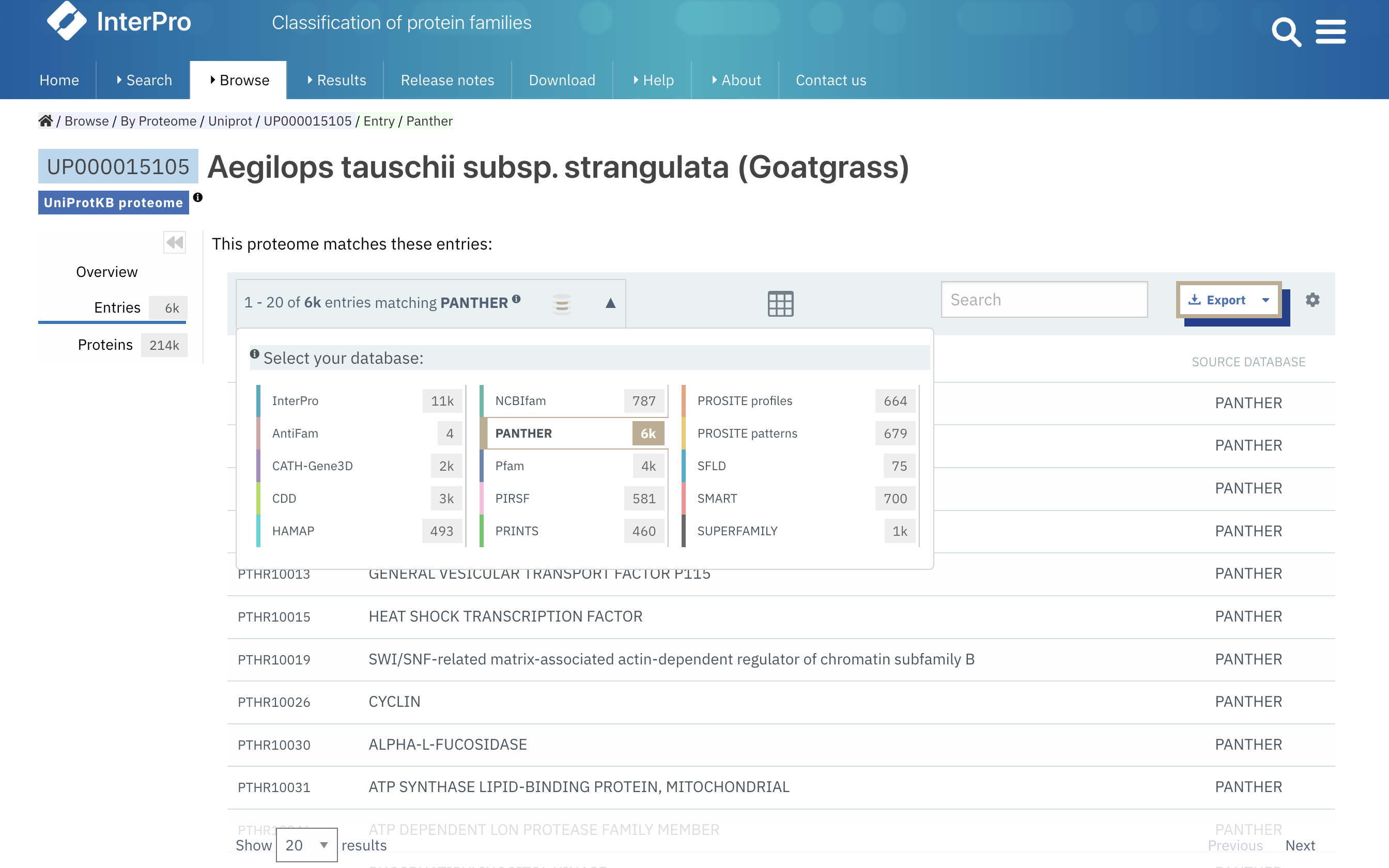 Figure 2. Selecting signatures from PANTHER matching the proteome UP000015105.
Figure 2. Selecting signatures from PANTHER matching the proteome UP000015105.
Link to AlphaFold prediction
We have added a link to access the AlphaFold prediction from the tables containing a list of proteins. For example, on the Proteins tab of an InterPro entry (Figure 3), a member database signature, or on the Similar Proteins tab in protein pages.
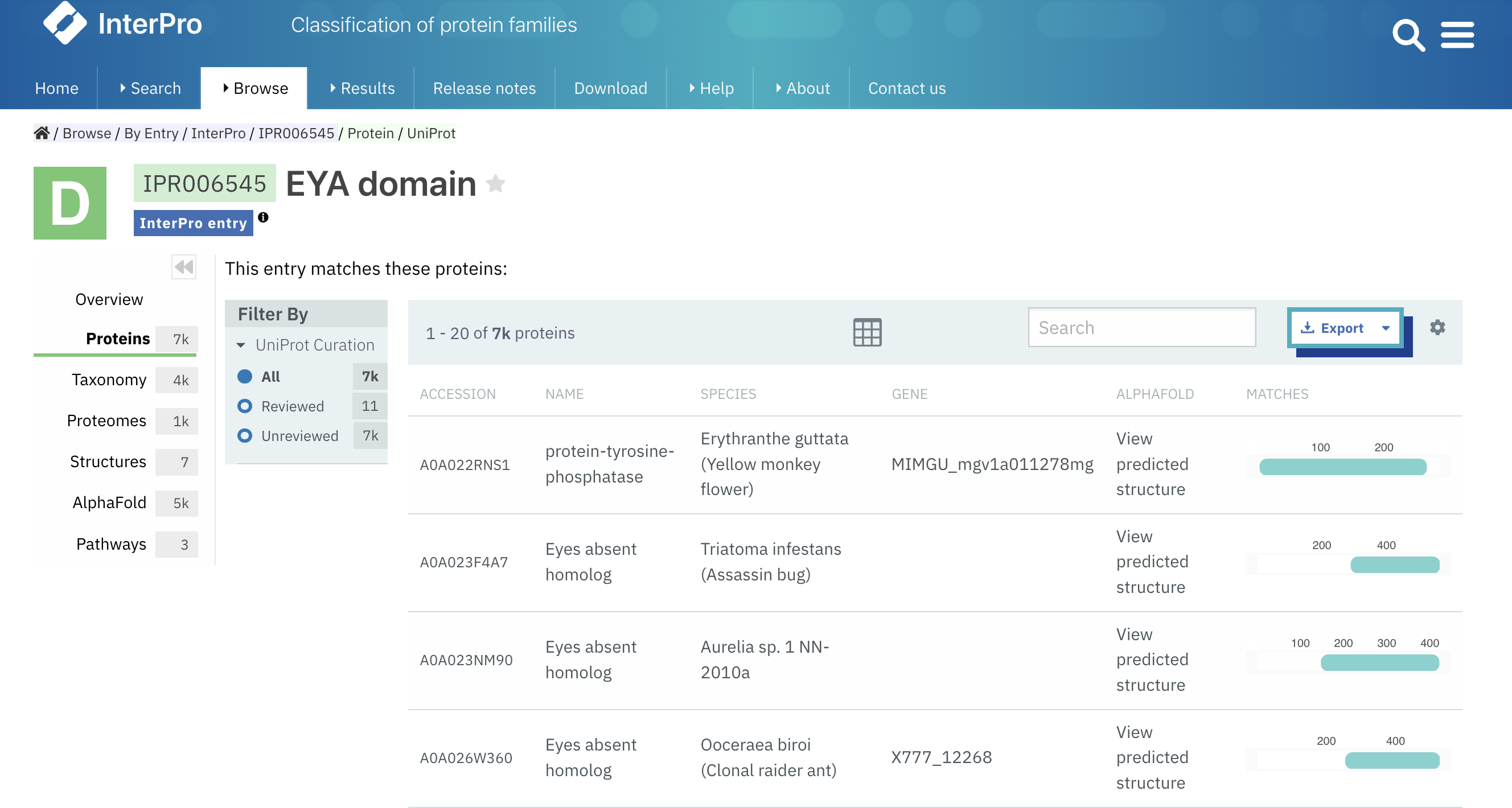 Figure 3. Example of a list of proteins with links to AlphaFold predicted structure (IPR006545).
Figure 3. Example of a list of proteins with links to AlphaFold predicted structure (IPR006545).
Addition of representative structures
We have added an image of a representative structure to the member database signature and InterPro entry pages, as shown in Figure 4. The chosen representative structure is picked from structures that match the entry and have a resolution of less than 2 Angstroms. In this refined dataset, the representative structure is identified as the one exhibiting the highest coverage ratio for the entry, where a minimum of 50% of the residues in the structure are covered by the entry.
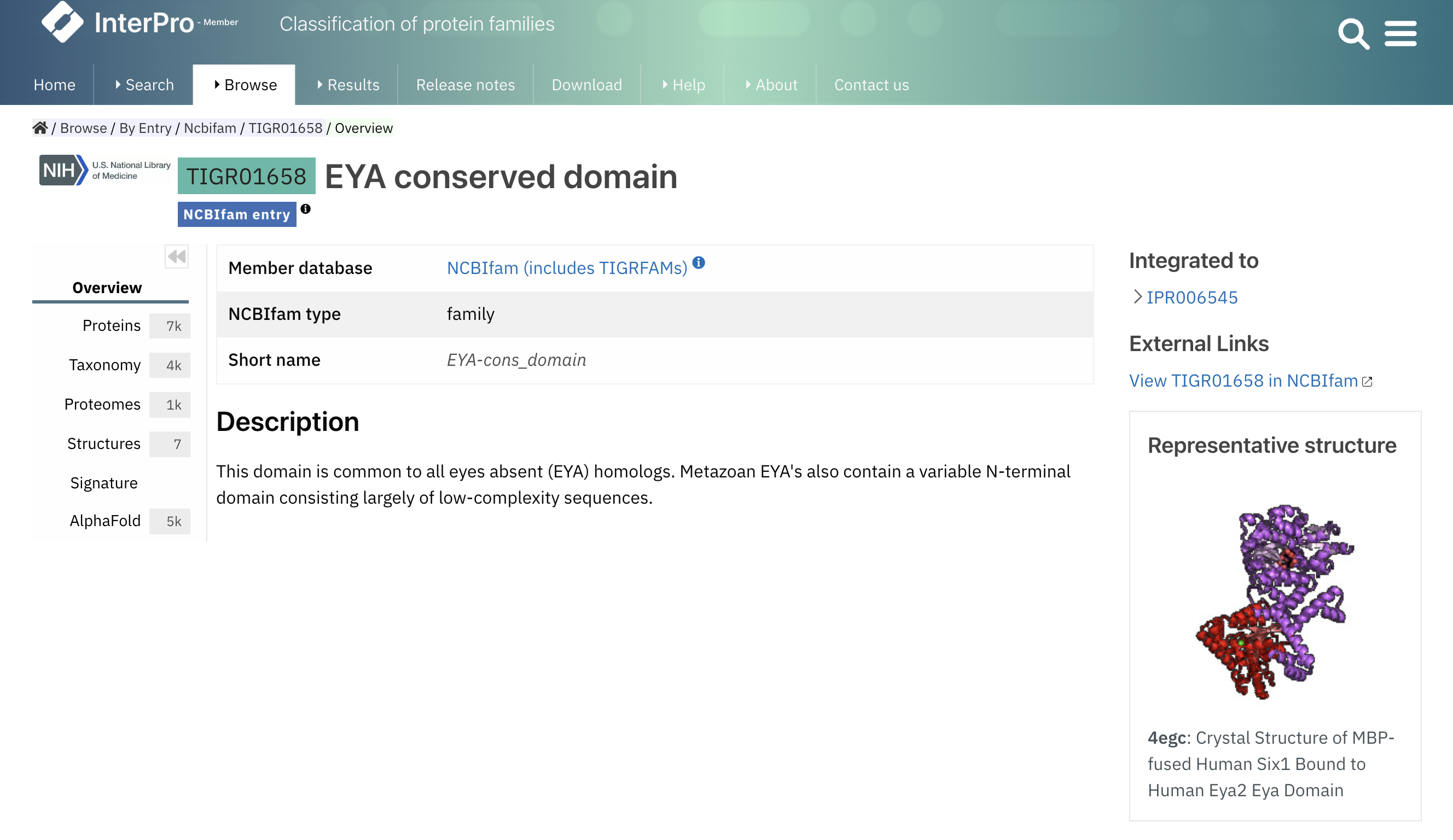 Figure 4. Representative structure for TIGR01658.
Figure 4. Representative structure for TIGR01658.
Citing Pfam
If you are using Pfam as part of your research, we’ll be glad if you could cite the latest Pfam paper. You can find a link to it and previous Pfam publications in the Help/Documentation section of the InterPro website menu.
