DNA Ligase IV
A family of Proteins
DNA ligases are a large family of proteins involved in DNA replication, repair and recombination. All DNA ligases follow the same reaction mechanism, but can be subdivided into two groups based on cofactor preferences: NAD+-dependent DNA ligases (EC 6.5.1.2; IPR0001679) are found only in eubacteria, while ATP-dependent DNA ligases (EC 6.5.1.1; IPR000977) are found in eukaryotes, archaea, eubacteria and viruses. In eukaryotes, three related classes of ATP-dependent enzymes (DNA ligases I, III and IV) are found, which differ in their ability to ligate a variety of nucleic acid substrates, and which have specific functions within the cell. DNA ligase I, which is conserved in all eukaryotes, is required during DNA replication for the ligation of Okazaki fragments during lagging-strand synthesis, as well as for DNA repair. DNA ligase III, which is unique to vertebrates, occurs as two distinct isoforms that arise by alternative splicing, one of which interacts with the mammalian DNA repair factor XRCC1. DNA ligase III functions to correct defective DNA strand-break repair, as well as in sister chromatid exchange during meiotic recombination.
DNA ligases I, III and IV are related in sequence and structure, being descended from a common ancestor. A series of motifs occur in the sequence alignments of ATP-dependent DNA ligases, which have been used to help construct the signatures in InterPro that define the family.
P49917 Human DNA ligase IV entry from InterPro
InterPro domain architecture
![]()
|
InterPro entry |
Method accession |
Graphical match |
Method name |
|
IPR000977 |
PF01068
|
|
DNA_ligase_A_M |
|
IPR000977 |
PF04675 |
|
DNA_ligase_A_N |
|
IPR000977 |
PF04679 |
|
DNA_ligase_A_C |
|
IPR000977 |
PS00333 |
|
DNA_LIGASE_A2 |
|
IPR000977 |
PS00697 |
|
DNA_LIGASE_A1 |
|
IPR000977 |
PS50160 |
|
DNA_LIGASE_A3 |
|
IPR000977 |
PS50161 |
|
DNA_LIGASE_A4 |
|
IPR000977 |
TIGR00574
|
|
dnl1 |
|
IPR001357 |
PF00533 |
|
BRCT |
|
IPR001357 |
PS50172 |
|
BRCT |
|
IPR001357 |
SM00292 |
|
BRCT |
|
IPR008994 |
SSF50249 |
|
Nucleic_acid_OB |
|
Classification |
PDB chain/Domain ID |
PDB chain/Structural Domains |
|
|
1ik9 |
1ik9c |
|
|
What InterPro Tells Us
From the graphical match table above, you can see that the signatures (method accession) are divided into three InterPro entries for human DNA ligase IV. These entries give information about the domain architecture of the protein, as well as its family relationships.
To look at the family
relationships that involve DNA ligase IV, we need to look at the entries at the
top of the table, IPR000977, which has 8 signatures
representing the ATP-dependent DNA ligase family. These signatures are: PF01068, PF04675 and PF04679 from the PFAM database, which
represent the ATP-dependent ligase domain, the N-terminal domain and the
C-terminal region, respectively, and which together detect members of this
family; PS00333, PS00697, PS50160 and PS50161 from the PROSITE database, which
represent the conserved regions found in the ATP-dependent DNA ligase family;
and TIGR00574 from the TIGRFAM database, which
covers the entire core of the protein family.
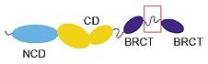
The domain architecture of DNA
ligase IV is represented in the ‘InterPro domain architecture’ (IDA) diagram at
the top of the page, as well as in the graphical match table. The IDA diagram describes two main domain
types: a DNA ligase domain and (two) BRCT domains. The DNA ligase domain is the central core of the enzyme that is
common to the ATP-dependent DNA ligase family, namely the N-terminal
non-catalytic region and the catalytic region together. The two BRCT domains are found only in some
DNA ligases, including DNA ligase IV, but are not common to the entire
family. A comparison of IDA with the
known domain architecture of DNA ligase IV is shown below:
|
|
|
|
The Graphical match table includes the signatures describing
some of these domains. Three signatures
represent IPR001357,
the C-terminal BRCT domain: PF00533 from the PFAM database, PS50172 from the PROSITE database, and SM00292 from the SMART database. IPR008994 is the nucleic acid-binding OB-fold
that forms part of the catalytic domain, and which is characteristic of many
nucleic-acid-binding protein; it is represented by one signature, SSF50249 from the SUPERFAMILY
database.
The remaining entry in the table is from the structural database PDB, and displays the region of the protein that has structural information (the name, 1ik9c, refers to PDB entry 1ik9, chain C). The region is very small, and corresponds to the insert region between the two BRCT domains, as depicted in the red-boxed region in the diagram above. This region is structurally important, because it is the site of binding to the XRCC4 protein, which undergoes conformational changes that are required for the NHEJ process.
What the Structure Tell Us
There are structures of DNA ligase IV and other DNA ligases in the Protein Data Bank (PDB); in particular the insert region between the two BRCT domains mentioned above. A detailed description and visualisation of the structural features of DNA ligase IV can be found at the PDB 'Molecule of the Month' .
Next: Table of DNA Ligases and NHEJ Proteins
Previous: NHEJ, How It Works