Interpro 85.0 New Features And Bug Fixes
interproThe latest release of InterPro comes with a number of improvements to the website. The list of changes is detailed below. If you have any feedback or suggestions, please contact us.
New features
Structural model subpage
Structure models and contact maps have been created for many of the Pfam families that do not have a structure in the PDB. They are available under the Structural model tab of Pfam signature pages. The models are generated using the automated trRosetta modeling pipeline developed by the Baker group. The contacts predictions between the sequence residues are shown at the top of the alignment and are highlighted in the 3D viewer when hovering over them as shown in Figure 1.
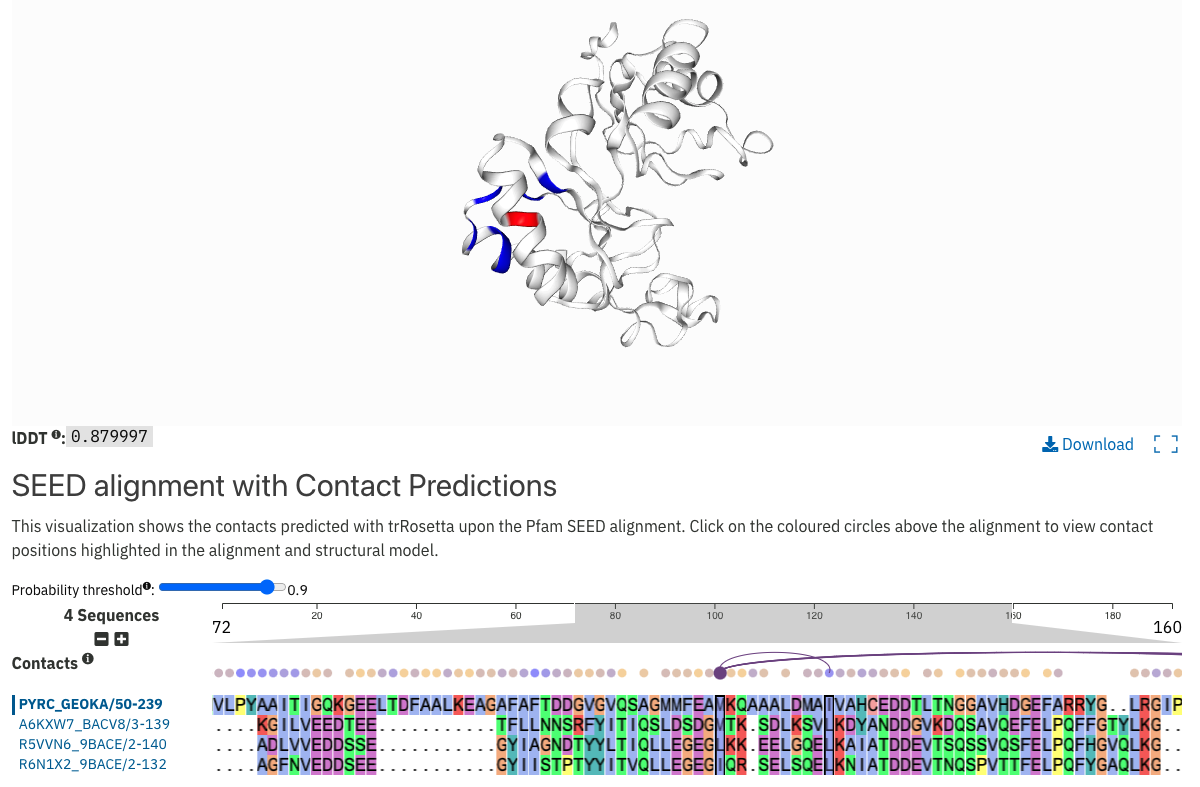 Figure 1. Contact map and structure prediction for Pfam entry PF12890.
Figure 1. Contact map and structure prediction for Pfam entry PF12890.
Updated conservation track
On protein pages the protein sequence viewer offers the possibility to visualise the amino acid conservation on the scale varying from 0 (not conserved) to 10 (highly conserved). Previously this information was calculated based on Pfam signatures, however Pfam entries don’t necessarily match the full sequence length. Now the calculation is based on PANTHER signatures ensuring a better coverage of the protein as shown in Figure 2 below. We also have changed the conservation representation to use a graph line instead of the heat map previously used.
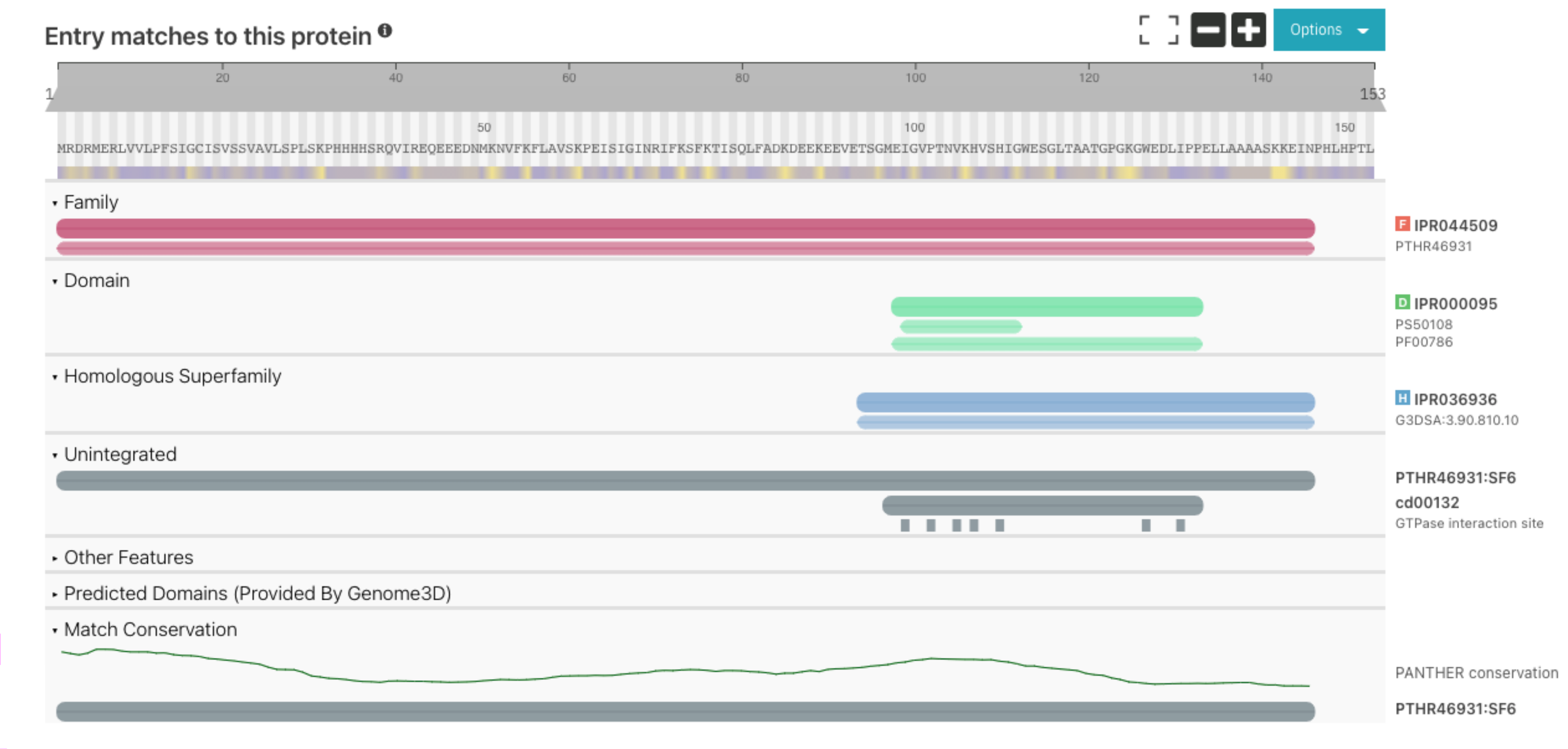 Figure 2. Conservation track old (A) vs new (B) display for UniProt Q9FFD5.
Figure 2. Conservation track old (A) vs new (B) display for UniProt Q9FFD5.
Scientific name search in Taxonomy tree
In the previous InterPro release, we have introduced an improved Scientific name search in the taxonomy table. In this release the search has been extended to the taxonomy tree view, allowing easier access to lower levels of the taxonomy without having to click on every node as shown in Figure 3.
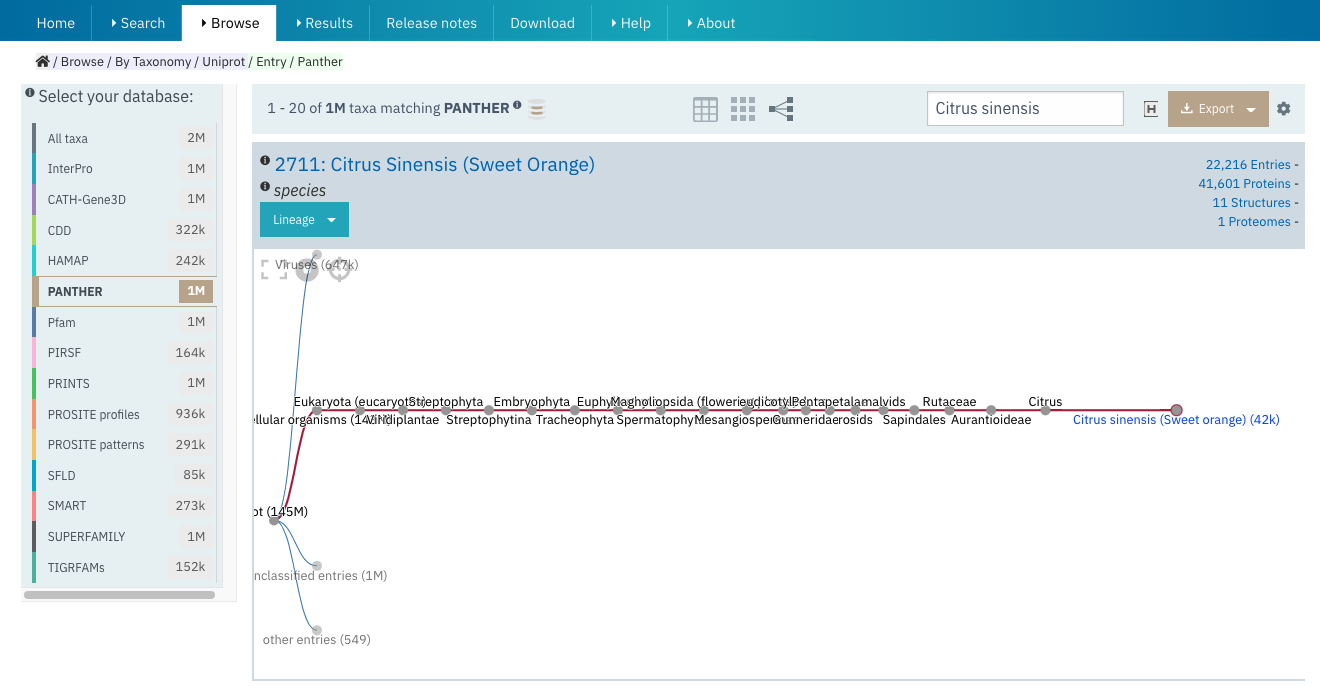 Figure 3. Scientific name search in taxonomy tree view for Citrus sinensis in InterPro Browse.
Figure 3. Scientific name search in taxonomy tree view for Citrus sinensis in InterPro Browse.
Fixes
Text search paste on mouse right click
With the recent change in the autocomplete library used in the text search, ‘paste on right click’ was not efficient. It has been fixed to allow users to paste when the mouse click is made within the search box.
Page footer overlapping Browse feature filters
There was an issue with the page footer overlapping with the Browse filters “Sequence status” when browsing All proteins, this is no longer the case.
Other fixes
Some links in InterPro entries descriptions pointing to UniProt accessions where not redirected to the correct InterPro page for unreviewed proteins, this has been resolved.
There was an issue highlighting some entries in the 3D viewer in the structure page, this has been fixed.
There is no more duplication of the protein viewer’s navigation track when choosing the print option in the sequence viewer.
FTP links for InterProScan have been replaced by https as major browsers are no longer providing FTP support.
The broken link of DOI article in the Home page is fixed.
