Interpro 100: A Special Release
interproThis month we are celebrating the InterPro centenary release! It includes new features and improvements to the InterPro website. The list of changes is detailed in this post. If you have any feedback or suggestions, please contact us.
Post content
- Data updates
- Search by protein name
- Short name shown in table of entries
- Links to Foldseek
- Pathogenic variants
- Updates to AI-generated tags
- Support of multiple Wikipedia pages
- EMBL-EBI survey
Data updates
In this release, we have updated Pfam to version 37.0 and updated InterPro entries accordingly. For more information about the Pfam 37.0 release, head to this blog post. We also have integrated 582 new signatures from the NCBIfam (26), Pfam (545), PANTHER (3) and CDD (8).
We have also updated Pfam-N data, available in the Other features section of the protein sequence viewer. This update uses Vision-Transformer Deep Learning Techniques, a different methodology compared to previous versions. The model has been trained using all InterPro member database signatures, including Pfam 36.0. Mode information about the model and its performances can be found in this Pfam blog post.
New features
Search by protein name
When looking at a list of proteins on the InterPro website, the list can be searched by UniProt accession, UniProt ID and protein name using the text search located on the table header (Figure 1). Please note that special characters other than letters and digits are not supported and should be removed from the protein name for the search to be successful.
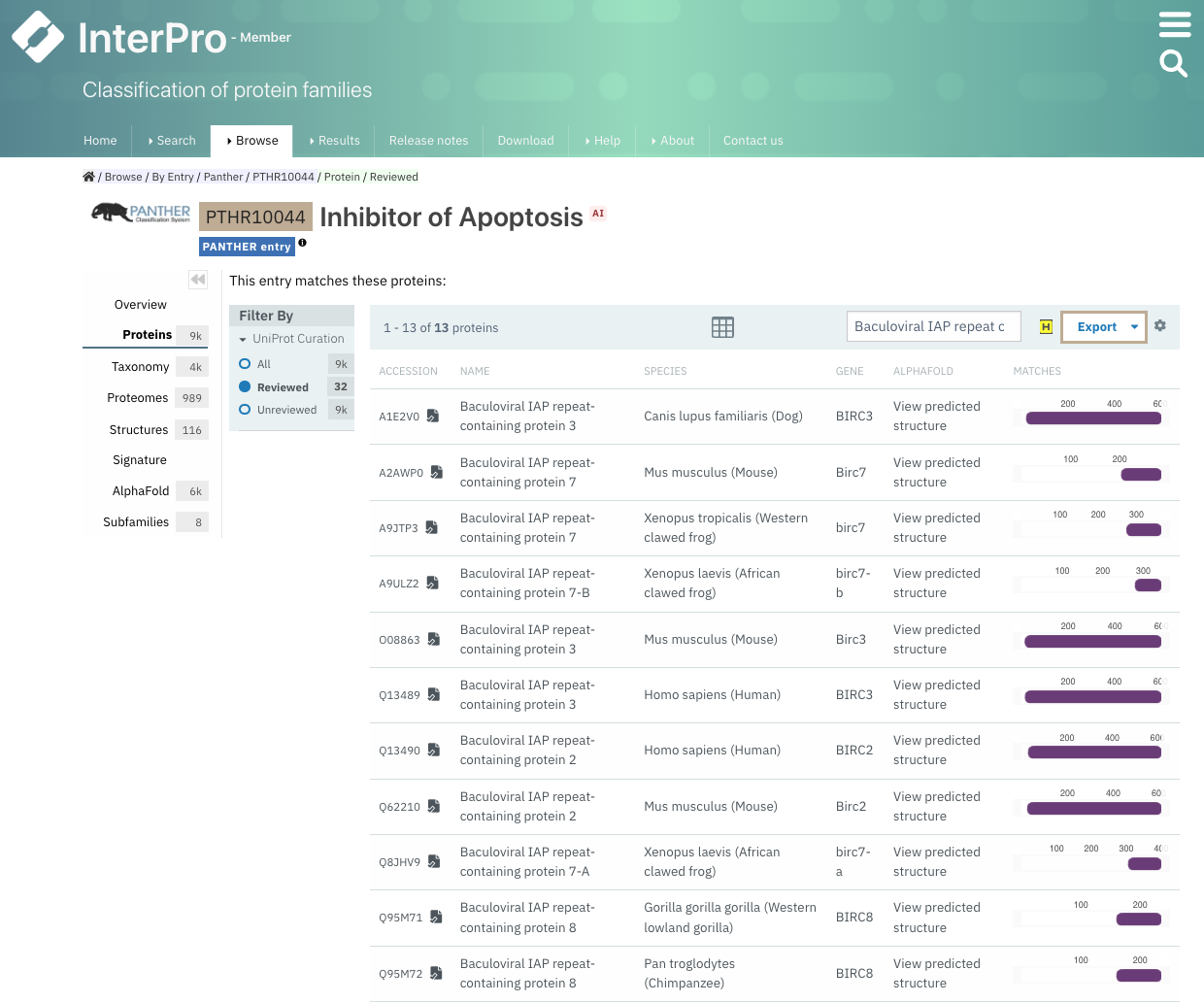 Figure 1. Search for Baculoviral IAP repeat containing protein in PANTHER PTHR10044 matches.
Figure 1. Search for Baculoviral IAP repeat containing protein in PANTHER PTHR10044 matches.
Short name shown in table of entries
For tables containing a list of InterPro entries or member database signatures, the following information is available: Entry accession, short name and name.
Links to Foldseek
The Foldseek Server offers fast and sensitive protein structure comparison search against large protein structure resources. A link to Foldseek has been added to AlphaFold and structure subpages, allowing search for proteins and PDB entries with similar 3D structures.
Pathogenic variants
We have added a new track to the protein sequence viewer to provide information about pathogenic and likely pathogenic residue variants in protein sequences (Figure 2). Please note that the data can differ from the data provided on the UniProt website as we are applying a Clinical significance filter, which is different from the one used by UniProt.
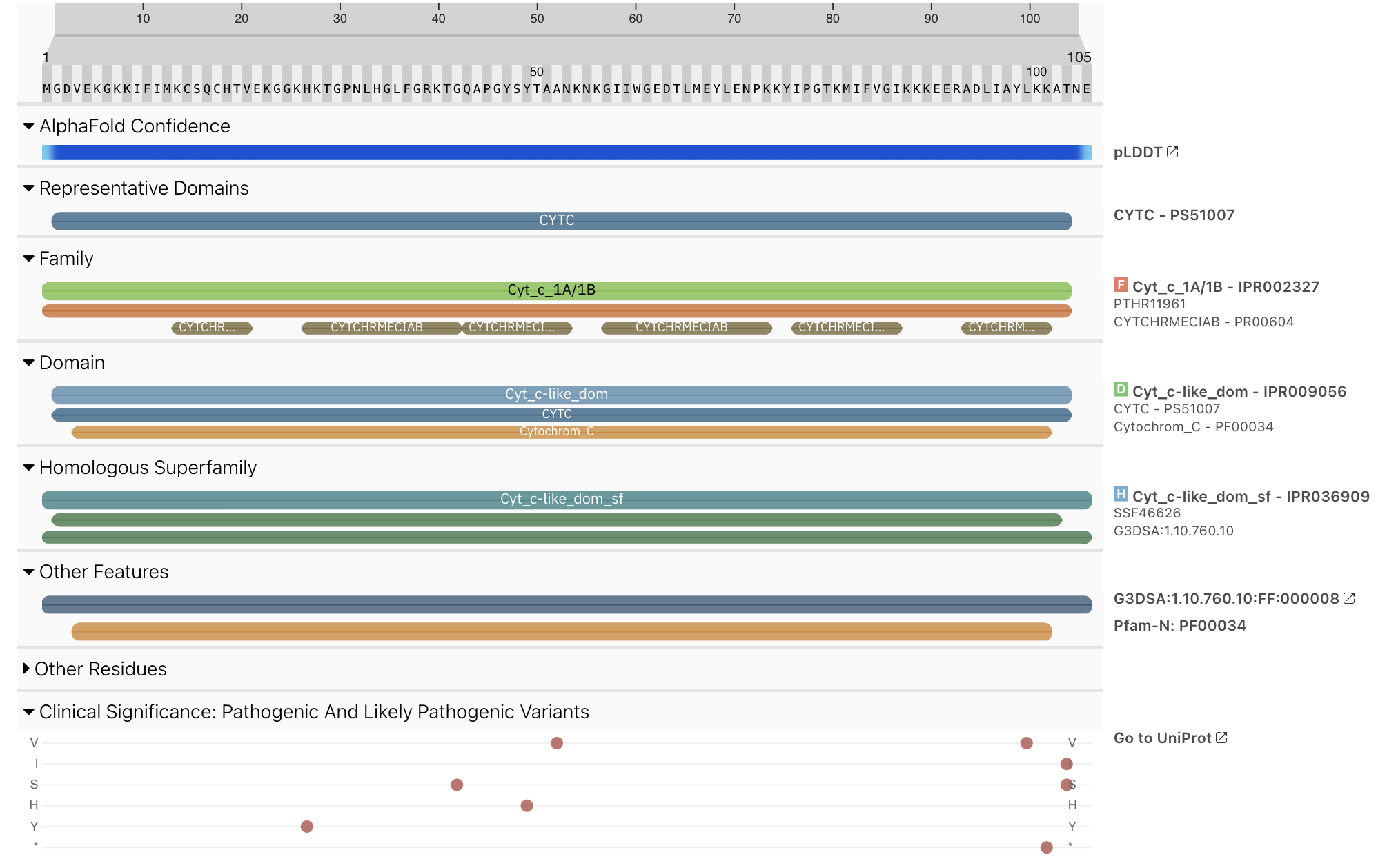 Figure 2. Example of pathogenic variant data for P99999.
Figure 2. Example of pathogenic variant data for P99999.
Updates to AI-generated tags
As previously mentioned, InterPro contains AI-generated entries. The InterPro data curation team may review these entries in the future. When that occurs, the curators may verify, accept, or amend the AI-generated content. The AI status tag can be the following:
- Unreviewed: the entry hasn’t been verified by a curator
- Reviewed: the entry has been verified by a curator
- Reviewed and updated: the entry has been verified and updated by a curator
The AI status is highlighted on the entry page in the tooltip when hovering over the AI symbol next to the entry name and short name, and by the AI tag above the entry descriptions (Figure 3).
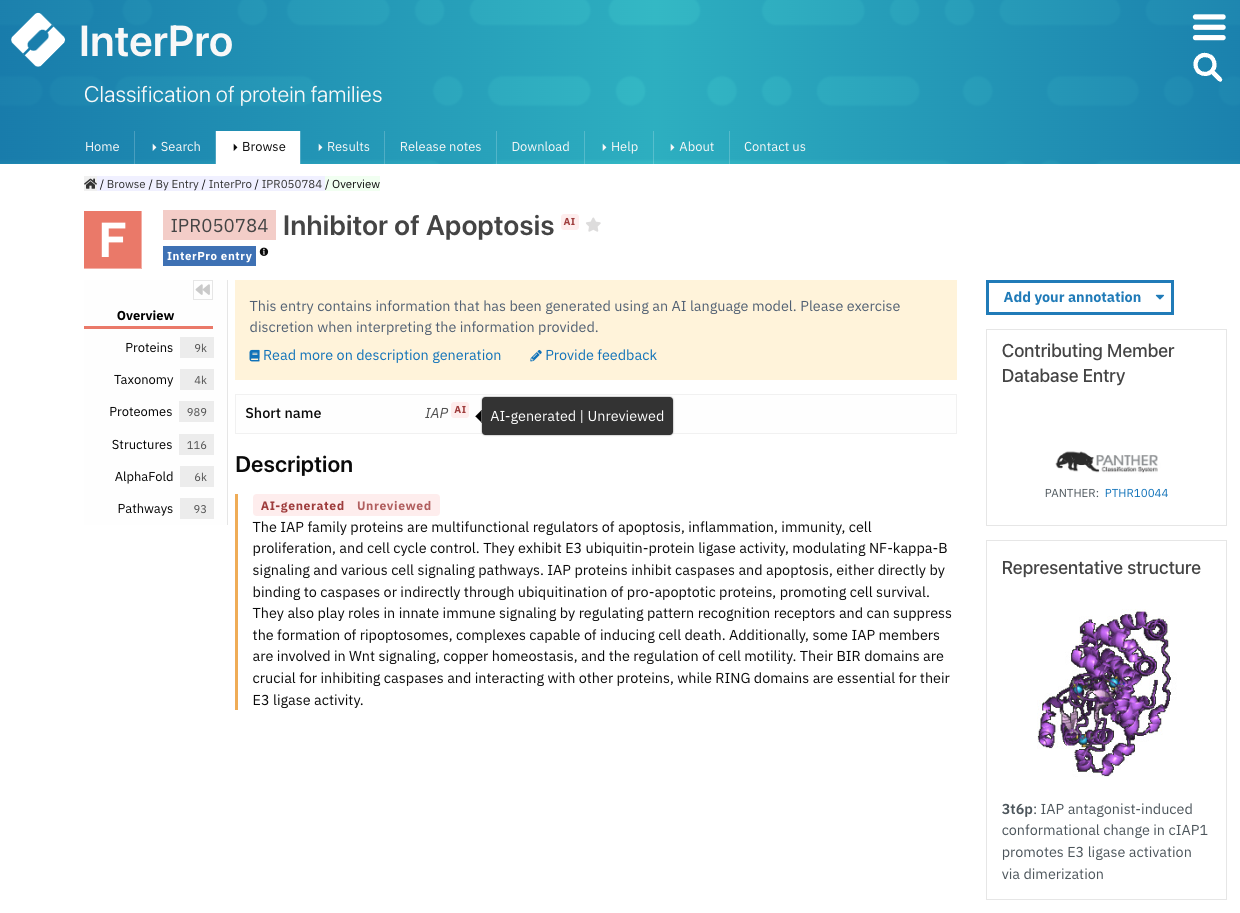 Figure 3. Example of an InterPro entry with an unreviewed AI-generated name, short name and description (IPR050784).
Figure 3. Example of an InterPro entry with an unreviewed AI-generated name, short name and description (IPR050784).
Support of multiple Wikipedia pages
Pfam encourages the annotation of Pfam entries via Wikipedia. Below the traditional description of the Pfam entry in the Overview tab of the Pfam family page, you may find one or more Wikipedia articles that provide a good description of the Pfam entry. Clicking on the title of the Wikipedia article opens the full article in a new tab.
For example, PF00001, 7 transmembrane receptor contains G-protein-coupled receptors (GPCR) which are a subgroup of the Rhodopsin-like receptors. Hence, two Wikipedia articles are linked to this entry (Figure 4).
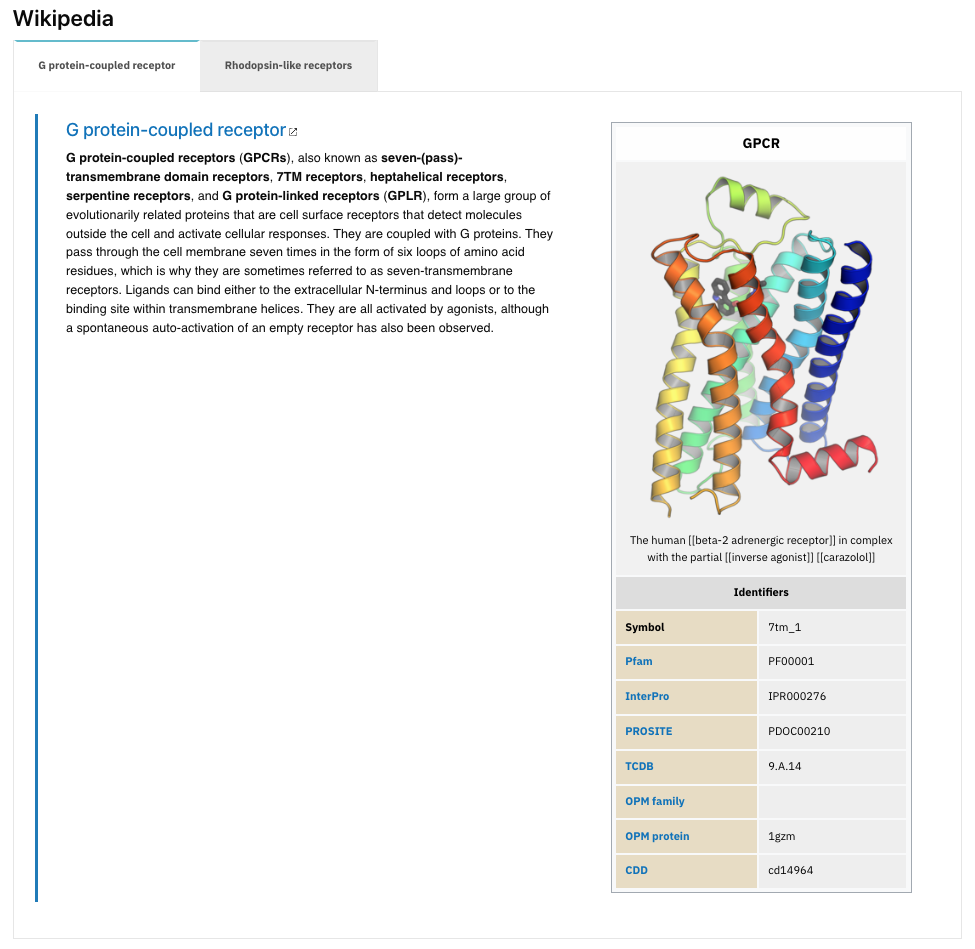 Figure 4. Example of the Wikipedia section with two articles associated with PF00001.
Figure 4. Example of the Wikipedia section with two articles associated with PF00001.
EMBL-EBI survey
Please spare a few minutes to tell us about your experience. By completing a short 10 minute survey, you’ll help us understand usage patterns, impact, and how we can improve our open data resources so they’re more useful to you.
Help shape the future of open data resources by sharing your experience.
The survey is open until June 17th, and we welcome responses from all over the world, regardless of usage level.
EMBL-EBI and collaborators manage over 40 open data resources spanning different data types for the life sciences. We rely heavily on the feedback of our user communities to shape the development of these data resources, and ensure they fulfil the ever-changing needs of our users. This is your chance to have a say.
