Interpro 95.0: New Features And Updates
interproThis release includes new features and improvements to the InterPro website. The list of changes is detailed below. If you have any feedback or suggestions, please contact us.
Content
In this release, we have updated NCBIfam to version 12.0 and updated InterPro entries accordingly. We also have integrated 424 new signatures from the NCBIfam (259), Pfam (2), PANTHER (5), CDD (157), SFLD (1) databases.
New resources
DisProt
DisProt is a database of intrinsically disordered proteins, where disordered regions are manually curated from the literature. We are providing Disprot annotations in InterPro, these annotations can be seen in the External Sources track in the protein sequence viewer when looking at a protein page.
RepeatsDB
RepeatsDB is a database of annotated tandem repeat protein structures. We are providing RepeatsDB annotations in InterPro, these annotations can be seen in the External Sources track in the protein sequence viewer when looking at a protein page.
ELM
The Eukaryotic Linear Motif (ELM) resource focuses on annotation and detection of eukaryotic linear motifs (ELMs) by providing both a repository of annotated motif data and an exploratory tool for motif prediction. ELMs, or short linear motifs (SLiMs), are compact protein interaction sites composed of short stretches of adjacent amino acids. They are enriched in intrinsically disordered regions of the proteome and provide a wide range of functionality to proteins. They play crucial roles in cell regulation and are also of clinical importance, as aberrant SLiM function has been associated with several diseases and SLiM mimics are often used by pathogens to manipulate their hosts’ cellular machinery (Davey, 2011, Uyar, 2014). We are now providing ELM annotations in InterPro. When a protein has an ELM annotation, it can be found in the Other Features track of the protein sequence viewer.
Protein sequence viewer updates
External Sources track
We have added a new track in the protein sequence viewer: External Sources. We have moved Genome3D annotations to be displayed in this category, additionally to DisProt and RepeatsDB, as shown in the figure below.
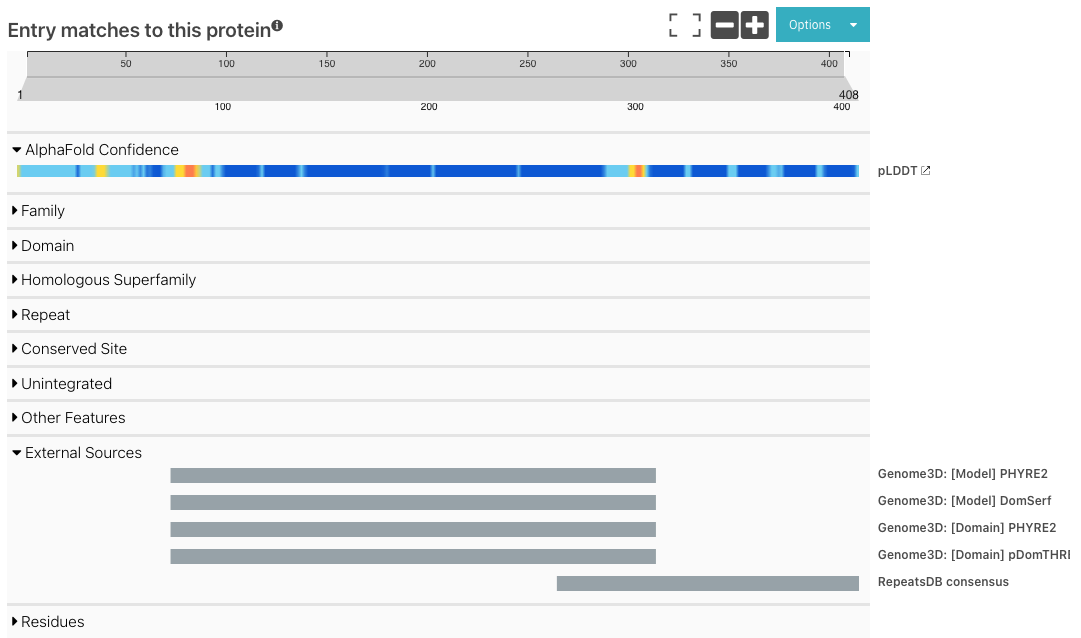 Figure 1. External Sources track for P63005, showing Genome3D and RepeatsDB annotations.
Figure 1. External Sources track for P63005, showing Genome3D and RepeatsDB annotations.
PANTHER GO terms
As previously announced, we have been providing PANTHER GO terms on protein pages, whenever available. These GO terms are now also available as part of the InterProScan results when performing a sequence search. Additionally, we now only display this category when GO terms are available.
Conservation track
The conservation track was previously available for proteins and was allowing the users to see the sequence conservation based on PANTHER models. However, due to the instability of the HMMER web server (https://www.ebi.ac.uk/Tools/hmmer), the service used to calculate the conservation, we had to disable this feature for the foreseeable future.
Fixes
As we are adding more and more annotations from various resources to be displayed in the protein sequence viewer, when enabling the full screen mode, some annotations were not visible. To resolve this issue, we have made the fullscreen view of the protein viewer scrollable.
Some PIRSR residues don’t have information about their location, we are now handling these cases.
New features
As introduced in InterPro 94.0, when a signature doesn’t have a description and is integrated in an InterPro entry, we import the description and references from the latter. When it is the case, we had added a tag when a citation was taken from the integrated signature in the description, this is now emphasised for the references section as well.
AlphaFold model mismatch
AlphaFold predictions are built using the sequences in UniProt. However, UniProt entries can be deleted or updated. For the former, we won’t display the AlphaFold prediction, because we no longer show the UniProt protein in InterPro. But for the latter, it might be an issue, especially if it is the protein sequence that has been updated. We have added a new feature which checks and displays an alert message if the sequence used in an AlphaFold model doesn’t match the current sequence available in UniProt, as shown in Figure 2.
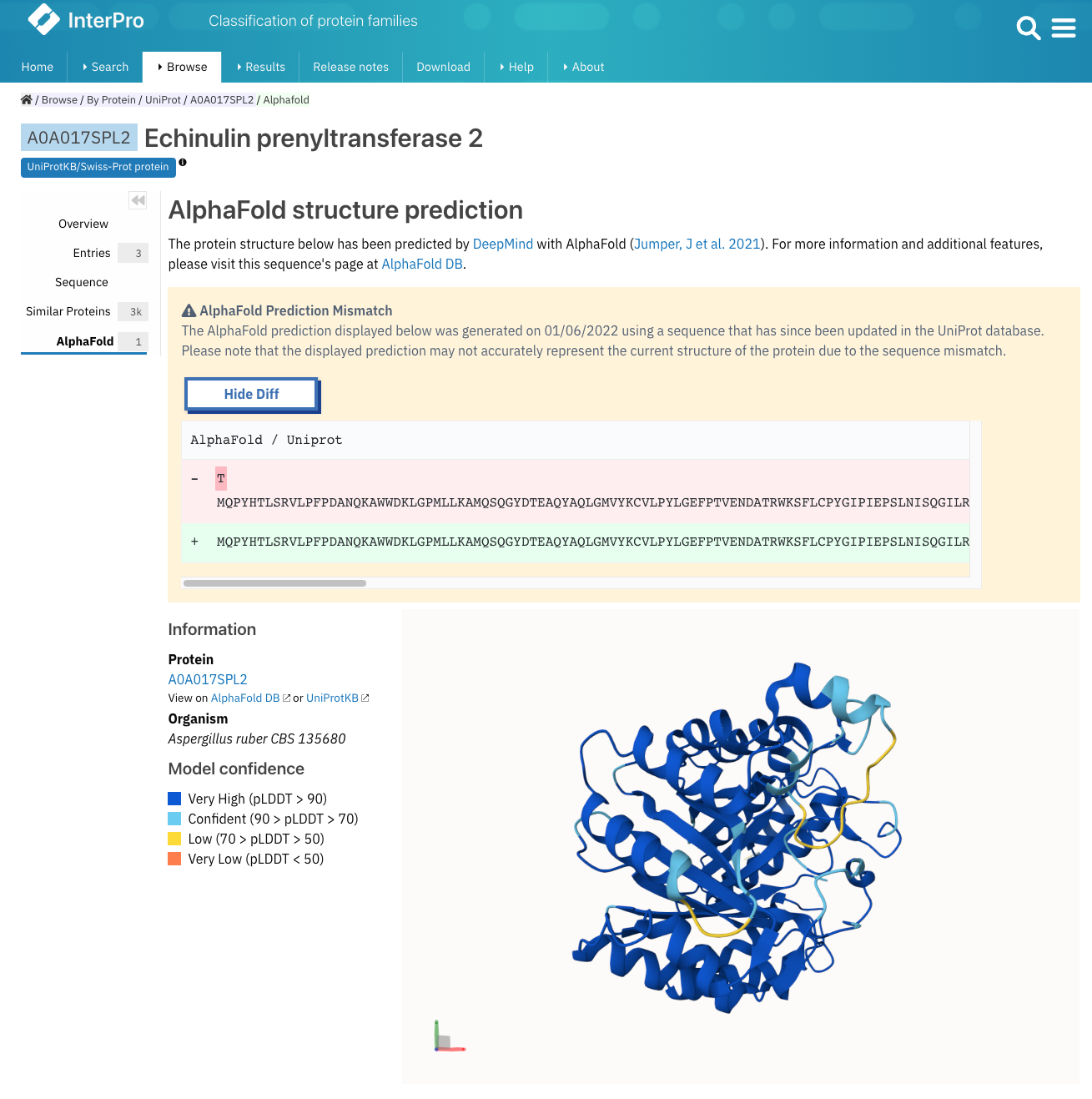 Figure 2. Example of an AlphaFold model where the UniProt sequence has changed (A0A017SPL2).
Figure 2. Example of an AlphaFold model where the UniProt sequence has changed (A0A017SPL2).
Migration of the InterPro codebase to TypeScript and Visual framework 2.0
As mentioned in our previous blog post, we have started the process to move the InterPro website code base to use TypeScript instead of JavaScript+FlowJS, and we are also migrating the codebase from using the EBI visual framework 1.4 (with foundation) to using Visual framework 2.0 (Vanilla CSS). In this release, we have migrated the following pages and components:
- The entry and signature pages title. We have also refactored it to display the accession first, additionally, and we have added a link to the member database website when clicking on the logo. And on NCBIfam entry pages we have added a tag saying: Includes TIGRFAMs.
- The Download page and components. Furthermore, we have added an extra tab containing the Antifam release files.
- The AlphaFold model subpage that is found in protein, InterPro entry and signature pages.
- The Panther GO terms displayed below the protein viewer in the Protein page and in the results of the sequence search.
- The different components related to the protein sequence viewer (e.g. Options, Tooltips and Labels)
- The Protein Overview subpage, the landing page displayed when looking at a protein page. The Isoforms component has also been migrated. Additionally, we have disabled the “HMMER search” option, due to the HMMER web server being unreliable at the moment.
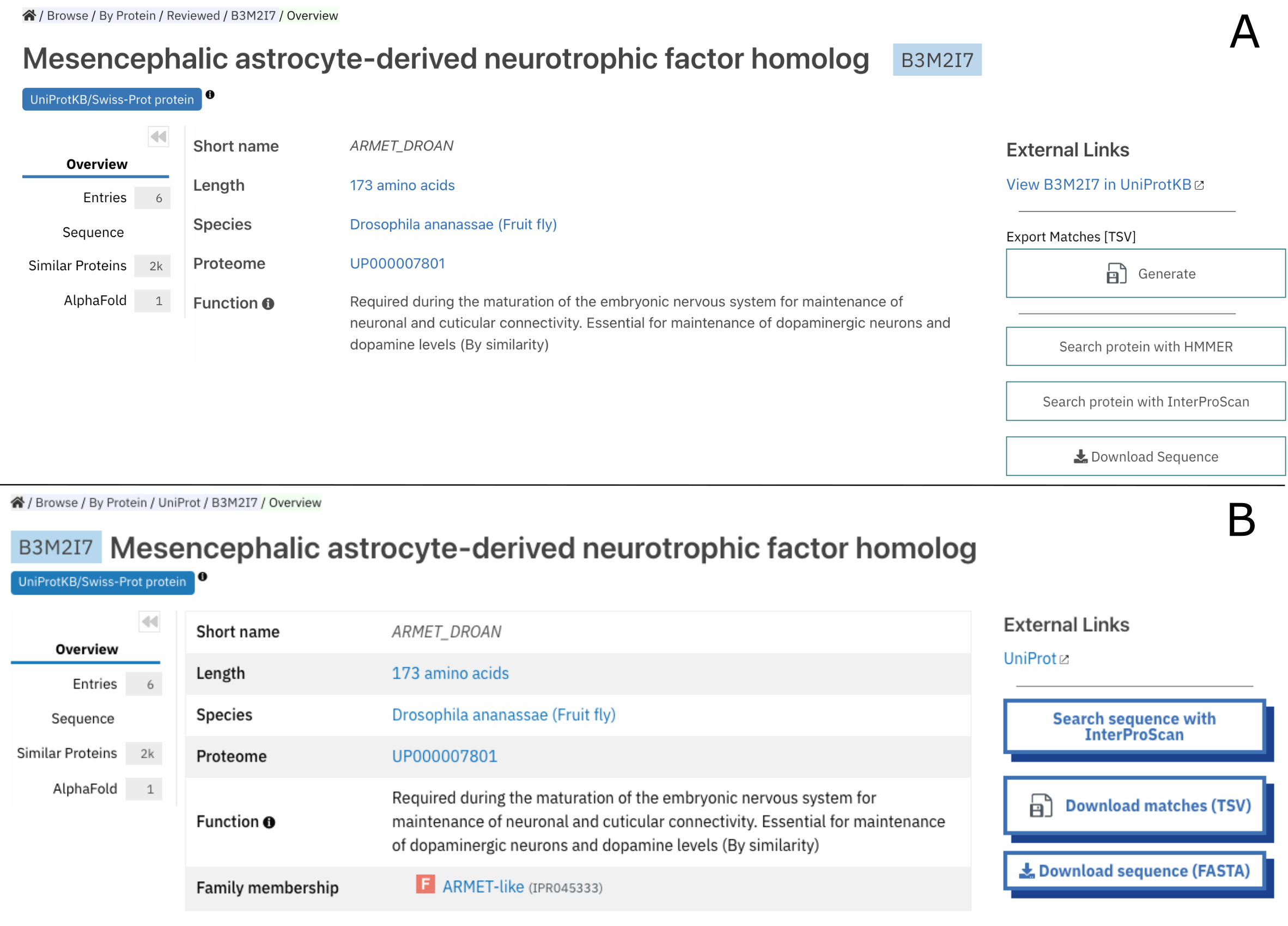 Figure 3. Protein B2M2I7 page before (A) and after (B) the update to Visual framework 2.0.
Figure 3. Protein B2M2I7 page before (A) and after (B) the update to Visual framework 2.0.
Fixes
On Set pages, we display a graphical view of the different signatures included in the set. The overlay on this viewer was causing a running time error, this issue has been fixed.
An issue had been reported informing us that the link to AlphaFold website in protein pages was not pointing to the right place, we have fixed it.
The restart download button, when a new version of the release is available, in Downloads (https://www.ebi.ac.uk/interpro/result/download/#table) couldn’t be clicked, as the pop disappeared straight away when the mouse was moved, this problem has been resolved.
The height of some domain architecture tracks was continuously growing, this issue has been fixed.
